Research Article - (2022) Volume 13, Issue 1
A Computational Study of PADI4 Receptor for Identifying Novel Drug Target against Rheumatoid Arthritis
Tooba Qamar, Prekshi Garg and Prachi Srivastava*Abstract
Background and objectives: Rheumatoid arthritis is an autoimmune disorder in which the immune system of the body loses the capability to differentiate between self and non-self cells. Patients with this disorder have autoantibodies like anti-citrullinated peptide antibody, which target their own healthy cells. PADI4 (Protein-Arginine Deiminase type-4) protein is found to be involved in pathophysiology of this disease, hence this protein was selected as a target for further studies. Till date only symptomatic treatments are available rather than permanent cure against it, hence research in such area is highly promising. The current in silico approach is based on identification of novel therapeutic inhibitors against PADI4 receptor using the computational researches.
Methods: In current study, the human Peptidyl Arginine Deiminase 4 (PADI4) receptor is the selected target in reference to designing of drug inhibitor against the Rheumatoid Arthritis. The 3D structure of the receptor was obtained from Protein Data Bank (PDB). A molecular docking study was performed between group of natural and synthetic compounds which possess proven anti-rheumatic activity with PADI4 receptor using Autodock tool 1.5.6.
Results: On the basis of parameters like lowest binding energy (kcal/Mol),hydrogen bond interaction, conformational structure, the three compounds namely Hydroxyanthroquinone(-7.40 kcal/mol), thymol (-5.97 kcal/mol), and vallinic acid (-5.90 kcal/mol) were screened out that shows very strong binding affinity with the active site of PADI4 receptor.
Interpretation and conclusion: The study concludes the high potential of hydroxyanthroquinone, thymol and vallinic acid to be active inhibitors against the PADI4 receptor protein. Thus, the molecular docking study will help in better understanding of the molecular interaction between ligand and receptor binding site thereby leading to development of novel therapeutics that can be considered as a prophylactic approach against this disease.
Keywords
Pharmaceutical industry, Public health, Legal framework
Introduction
Rheumatoid Arthritis (RA) is a chronic, inflammatory autoimmune disorder in which the immune system of the body targets its own healthy cells. Such individual have unusual autoantibodies like anti-carbamylated protein antibodies, Anti-Citrullinated Protein Antibody (ACPA) (Spengler J, et al., 2015) and Rheumatoid Factor (RF) that keeps on circulating in their blood resulting into the target of their own body tissues, as these antibodies lack the ability to distinguish between self and non-self cells. RA initially targets small joints, later on it affects larger joints resulting into the weakening of tendons and ligaments. All these damage eventually results into bone erosion and painful joint inflammation (Boissier MC, et al., 2012). Some of the clinical symptoms are fever, fatigue, joint stiffness, and formation of swollen rheumatoid nodules, painful and inflamed joints. One of the important hallmark of RA is the development of hyperplasia of the synovial membrane (Moussaieff A, et al., 2007). If this disease is not detected and treated at initial stage, then it can lead to complete joint destruction along with cardiovascular and renal manifestations.
Rheumatoid arthritis, being a multifactorial disorder is caused by both genetic and environmental factors. Many polymorphic forms of the genes are found associated with this disease like HLA-DRB1 gene association with Protein tyrosine phosphatase which is a non-receptor type 22 (PTPN22) alleles found in ACPA antibodies. The rate of morbidity of RA in women is 2-3 times higher as compared to men because it was found that women have high percentage of CD4+ T cells, and Th1 cytokine than men that results into an up regulation of CD4/CD8 ratio (Auger I, et al., 2012). The dysregulation of estrogens, prolactin and hormones contributes towards immunopathogenesis of RA. The immunopathogenesis of RA involves the dysfunctioning of innate immunity, adaptive responses against self-antigens, immune-complex mediated complement activation, imbalance in cytokine networks and immune regulation (Burmester GR and Pope JE, 2017).
Peptidylarginine deiminase (PADI) enzyme five different isozymes namely PADI1, PADI2, PADI3, PADI4 and PADI6. Since PAD4 is most stable isoform as compare to other, so their increase expression and function was found to be associated with risk of Rheumatoid Arthritis. Amongst them, PADI4 is a calcium dependent homodimeric enzyme with 663 amino acid residues that is responsible for catalyzing the hydrolysis of peptidyl arginine residues to peptidyl citrulline via deimination in the presence of divalent calcium ions (Ca2+) known as citrullination (Kourilovitch M, et al., 2014). It is encoded by gene PADI4. Some preferred susbstrates of PADI4 for citrullination are histones H3 and H4 at various sites of Arginine residues. Generally these proteins over citrullinate arginine 3 residue of histone H4 and arginine 17 residue of histone H3 along with non-histone protein vimentin of macrophage's, resulting into proliferation of Fibroblast-like Synoviocytes (FLS), degeneration of cytoskeletal and thereby forming rheumatoid pannus (Smolen J, et al., 2016). These proteins are known to play a vital role in immunopathogenesis of RA and show their dominance in synovial tissue. In case of RA affected individuals, these proteins are overexpressed in synovial organization and Peripheral Blood Mononuclear Cells (PBMCs). Therefore, abnormal expression of PADI4 is a key biomarker for disease susceptibility in RA. The dysregulated mechanism of PAD4 protein at molecular level in RA is found to be linked with onset and progression of RA, resulting into hyperplasia and complete synovial joint destruction (Turunen S, et al., 2016).
Since PADI4 protein is found to be involved in pathophysiology of this disease and causes inflammation of joints hence this protein was selected as a target for further studies. Till date only symptomatic treatments are available, there is no permanent cure for this disease. The current in silico approach was done to identify a novel therapeutic inhibitor against PADI4 receptor using the computational drug discovery based pipeline (Baji P, et al., 2014). These natural inhibitors can be considered as a prophylactic approach against RA as the available DMARDs have many shortcomings that restrict their use (Laurent L, et al., 2015). in silico research is continuing to play a vital role in treating these infectious maladies and is therefore having number of advantages too. One such benefit is that they employ various databases of the organic structures and quickly generate results with lost cost. They even don't need any facilities regarding the biosafety of the compounds. Results highlighting the efficacy of this type of work are numerous (Gilbert D, 2004). RA is declared as a global health problem and the annual disease susceptibility rate of this disease in the United States and many northern European countries is reported to be about 40 per 100,000 persons. In India, about 23 million people are affected annually. According to experts, they predicted that the number of patients with RA may double by 2030 (Koushik S, et al., 2017). As per the latest research, there is no permanent cure for this disease, only the symptoms can be controlled and affected individual have to rely on Disease-Modifying Antirheumatic Drugs (DMARDs) like methotrexate, leflumomide, etc. These DMARDs are not much preferred because they are costly ($450/year to $25,000/year) and are associated with several types of toxicities like renal and liver toxicity, allergic reactions neutropenia, pneumonia (Khalid Z and Sezerman OU, 2018). Therefore, there is an urgent need for the development of more effective and rational approach of drug designing that can be specific and have less toxicity issues against rheumatoid arthritis (Dudley ET, et al., 2011).
Methodology
Prediction of protein structure and validation
The 3D structure of the PADI4 receptor was retrieved from protein databank database (PDB) with accession ID-1WD9 and DOI-10.2210/pdb- 1WD9/pdb. The retrieved protein is human peptidylarginine deiminase type 4 (PADI4) which is calcium bounded and is classified as hydrolase and expression system is Escherichia coli BL21 (DE3). The X-ray diffraction technique was used to determine the protein structure (Vakser IA, 2014). The 3D structure was subjected to validation according to the percentile scores that ranges between 0-100 for global validation metrics of the PADI4 entry and is represented in the form of graph. The Ramachandran outliers determine the possibility of steric clashes and structural reliability associated with PADI4 (Moreira IS, et al., 2010).
Identification of ligand
The thirty natural compounds and five synthetic compounds which were proven in vitro and in vivo anti-rheumatic activity were utilized in the study based on different literature reviews. The virtual library of natural compounds (ligands) was made against rheumatoid arthritis as depicted in Table 1 and for synthetic compounds is shown in Table 2. The Pub Chem database was used to retrieve the available 2D and 3D structures of compounds. Further, analysis of molecular and bioactivity along with drug-likeness properties of the selected compounds were done by Molinspiration chemoinformatics software (Huang YF, et al., 2013). It allows easy visualization of different molecules that are encoded as SMILES or SD file and thereby supporting the calculation of vital molecular properties such as milogP, molecular weight, number of hydrogen bond donors and acceptors along with prediction of bioactivity score for the most crucial drug targets like kinase inhibitors, GPCR ligands, nuclear receptors (Miao Z and Westhof E, 2017).
| S. No | Compound name | Plant derived | Mol. wt (g/mol) | H-bond donor | H-bond Acceptor |
|---|---|---|---|---|---|
| 1 | Flavone | Vitex negundo | 222.24 | 0 | 2 |
| 2 | Thymol | Trachyspermum ammi | 150.22 | 1 | 1 |
| 3 | Nicotinic Acid | Aegle marmelo | 123.11 | 1 | 3 |
| 4 | Gingerol | Zingiber officinale | 294.39 | 2 | 4 |
| 5 | Glabrene | Glycyrrhiza glabra | 322.36 | 2 | 4 |
| 6 | Coumarins | Urtica pilulifera | 146.15 | 0 | 2 |
| 7 | Cinnamaldehyde | Cinnamomum verum | 132.16 | 0 | 1 |
| 8 | Saponins | Phrynium imbricatum | 1223.36 | 15 | 27 |
| 9 | Hydroxyanthraquinones | Justicia gendarussa | 224.22 | 1 | 3 |
| 10 | Eugenol | Hibiscus platanifolius | 164.2 | 1 | 2 |
| 11 | Curcumenol | Curcuma longa | 234.34 | 1 | 2 |
| 12 | Vanillic acid | Salix alba | 168.15 | 2 | 4 |
| 13 | Boswellic acid | Boswellia serrata | 456.71 | 2 | 3 |
| 14 | Salicyclic acid | Aloe barbadensis | 138.12 | 2 | 3 |
| 15 | Capsaicinoids | Capsicum frutescens | 305.42 | 2 | 4 |
| 16 | Charantosides | Cassytha filiformis | 630.86 | 4 | 8 |
| 17 | Thymol | Thymus vulgaris | 150.22 | 1 | 1 |
| 18 | Piperine | Piper nigrum | 285.34 | 0 | 4 |
| 19 | Terpinen-4-ol | Cinnamomum camphora | 154.25 | 1 | 1 |
| 20 | Caffeic acid | Borago officinalis | 180.16 | 3 | 4 |
| 21 | Cinnamic acid | Harpagophytum procumbens | 148.16 | 1 | 2 |
| 22 | Oleanic acid | Nyctanthes arbor tristis | 456.71 | 2 | 3 |
| 23 | Phloraspine | Curcuma longa | 432.47 | 5 | 8 |
| 24 | Alpha-thujone | Salvia miltiorrhiza | 152.24 | 0 | 1 |
| 25 | Menthol | Mentha arvensis | 156.27 | 1 | 1 |
| 26 | Triterpenoids | Aristolochia bracteata | 552.77 | 3 | 7 |
| 27 | Pannarin | Boerhaavia diffusa | 362.76 | 1 | 6 |
| 28 | Physcion | Caesalpinia sappan | 136.24 | 0 | 0 |
| 29 | Mannite | Punica granatum | 182.17 | 6 | 6 |
| 30-Jan | Epigallocatechin gallate | Camellia sinensis | 458.38 | 8 | 11 |
Table 1: Virtual library of natural compounds
| S. No | Compound name | Mol. wt (g/mol) | H-bond donor | H-bond acceptor |
|---|---|---|---|---|
| 1 | Methotrexate | 454.45 | 7 | 13 |
| 2 | Etoricoxib | 358.8 | 0 | 4 |
| 3 | Leflunomide | 270.21 | 1 | 4 |
| 4 | Hydroxychloroquine sulfate | 335.88 | 2 | 4 |
| 5 | Tofacitinib | 312.37 | 1 | 7 |
Table 2: List of synthetic compounds
Molecular docking study
Rigid molecular docking study was performed between the PADI4 receptor and ten natural and two synthetic compounds using the Autodock 1.5.6 tool to predict the best binding score, affinity, and confirmation (Pagadala NS, et al.,2017). AutoDock is an automated suite of protein-ligand docking tools used to predict the protein interactions with small molecules such as drug molecules and substrate. It analyzes the interaction of ligand molecules at the specified target site of the protein. AutoDock uses AMBER force field and linear regression to predict the free binding energies. The lowest binding energy was the criteria undertaken to select the best protein-ligand complex. Calculation of binding energy was performed using a semi-empirical free energy force field with charge-based desolvation and grid-based docking. The force field was selected based on a comprehensive thermodynamic model that allows the incorporation of intermolecular energies into the predicted binding energy (Morris GM, et al., 2009). Cygwin platform was used to run docking files. Discovery Studio 4.5 was used to visualize and analyze protein-ligand interaction and confirmation (Umeda N, et al., 2016).
Results and Discussion
3D structure prediction and validation
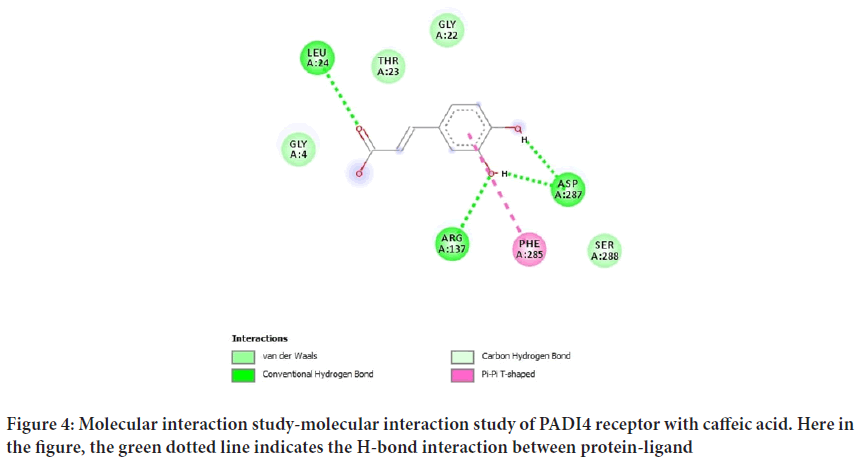
The total length of the PADI4 receptor in humans is 663 amino acids with a molecular weight of 75.27 kDa, atom count as 4829, modeled residue count of 603 and deposited residue count of 670 with one unique protein chain. It has a resolution of 2.60 Å with R-value free as 0.269, R-value work of 0.224 and R-value observed as 0.228. It is shown in Figure 1A. The percentile scores between 0-100 for global validation metrics of the PADI4 are depicted in Figure 1B. The depicts of the geometric issues that were reported across the polymeric chains and their respective fit to the electron density. The red, yellow, orange and green segments on the lower bar shows the fraction of residues that comprises of outliers for >=3, 2, 1 and 0 types of geometric quality criteria respectively whereas a grey segment depicts the fraction of residues that are not modeled (Bovac N, 2013).The numeric value for each and every fraction is shown below the corresponding segment, with the help of a dot representing fractions <=5%. The upper red bar shows the fraction of residues that have bad fit to the electron density.
Figure 1: Protein structure: Structure prediction and validation of PADI4 receptor. (A) 3D structure of PADI4 receptor (B) Ramachandran plot of PADI4 receptor
The geometric configurational values of PADI4 per unit cell is depicted in Table 3. Here, the crystal structure of receptor is the Ca2+ free wild-type PADI4, that exhibits that the polypeptide chain acquires an elongated fold with N-terminal domain forming two subdomains that are similar to immunoglobulin and the C-terminal domain that forms an alpha/beta propeller conformation (Arita K, et al., 2004). Moreover, it has five Ca2+ binding sites where none of these adopts an EF-hand motif (helix-loop- helix topology). The results of the structural data revealed that changes in conformation occur after binding of Ca2+ to the receptor (Kim S, et al., 2018). This results in generation of active site cleft. The geometric configurational values of PADI4 per unit cell are depicted in Table 4.
| Length (Å ) | Angle ( ° ) |
|---|---|
| a=146.389 | α=90 |
| b=60.094 | β=124.37 |
| c=115.339 | γ=90 |
Table 3: The geometric configurational values of PADI4 per unit cell
| S. No | Ligand | Binding energy (Kcal/Mol) |
|---|---|---|
| 1 | Cinnamic acid | -5.21 Kcal/Mol |
| 2. | Terpinen | -5.39 Kcal/Mol |
| 3. | Menthol | -5.87 Kcal/Mol |
| 4. | Caffeic acid | -5.45 Kcal/Mol |
| 5. | Hydroxyanthroquinone | -7.4 Kcal/Mol |
| 6. | Nicotinic acid | -5.02 Kcal/Mol |
| 7. | Vallinic acid | -5.9 Kcal/Mol |
| 8. | Thymol | -5.97 Kcal/Mol |
| 9. | Salicylic acid | -5.02 Kcal/Mol |
| 10. | Eugenol | -5.44 Kcal/Mol |
| 11. | Tofacitinib | -7.04 Kcal/Mol |
| 12. | Methrotrexate | -6.28 Kcal/Mol |
Table 4: Molecular docking results
| S. No | Ligand | Binding energy (Kcal/Mol) | H-bond interactions |
|---|---|---|---|
| 1 | Hydroxyanthroquinone | -7.4 Kcal/Mol | HIS-361, ILE-149, SER-181 |
| 2 | Thymol | -5.97 Kcal/Mol | HIS-361, ASP-120, |
| 3 | Vallinic acid | -5.9 Kcal/Mol | VAL-18, LEU-24,GLY-4,THR-23, THR-5, CYS-19 |
| 4 | Menthol | -5.87 Kcal/Mol | GLN-136 |
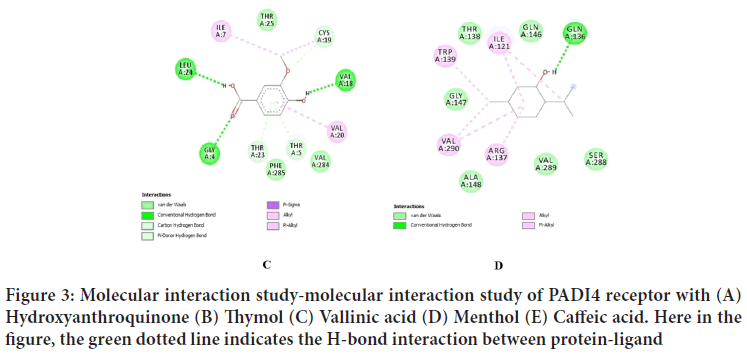
| 5 | Caffeic acid | -5.45 Kcal/Mol | ARG-137, ASP-287, LEU-24, SER-288 |
Table 5: Best five docking results
Molecular docking study
One of the best reliable methods for analyzing and predicting the best fit protein-ligand confirmation is molecular docking study (Savarino L, et al., 2005). In this study, molecular docking was performed between PADI4 receptor and thirty compounds using Autodock 1.5.6 tools. This was done in order to obtain a higher stable protein-ligand complex (Anderson AC, 2003). The results of molecular docking for top ten natural compounds and two synthetic compounds were depicted in Table 5 and for best five natural compounds are shown in Table 5. On the basis of parameters like lowest binding energy that is denoted by ΔGb (kcal/Mol), hydrogen bond interaction, conformational structure, the three compounds namely hydroxyanthroquinone (-7.40 kcal/mol) (Figure 2), thymol (-5.97 kcal/mol) (Figure 3), and vallinic acid (-5.90 kcal/mol) were screened out that shows very strong binding affinity with the active site of PADI4 receptor (Figure 4) (Luo H, et al., 2016 and Dong LM, et al., 2017). Thus, these compounds can be taken into consideration as a suitable therapeutic inhibitor against rheumatoid arthritis. Later, all these three dock complexes can be subjected to a molecular dynamics simulation study for checking the stability of protein-ligand complexes (Liu H, et al., 2016). Rheumatoid arthritis is dreadful systemic autoimmune disorder which is declared as a global health problem, adversely affecting approximately about 350 million people and is chronic in North America. The annual disease susceptibility rate of rheumatoid arthritis in the United States and many northern European countries is reported to be about 40 per 100,000 persons. In India, about 23 million people are affected annually. If specific and accurate treatment is not given at an initial stage, then it can lead to overall joint destruction leading to complete disability (Chen JJ, et al., 2011). As per the latest research, there is no permanent cure for this disease, only the symptoms can be controlled and affected individual have to rely on Disease-Modifying Anti-Rheumatic Drugs (DMARDs) like methotrexate, leflumomide, sulfasalazine (Kuo WL, et al., 2014). These DMARDs are not much preferred because they are costly ($450/year to $25,000/year) and are associated with several types of toxicities like renal and liver toxicity, allergic reactions neutropenia, pneumonia. Therefore, there is an urgent need for the development of more effective and rational approach of drug designing that can be specific and have less toxicity issues. In this regard, the computational in silico approach holds an excellent promising result in the discovery and development of drug with cost effectiveness (Okimoto N, et al., 2009). In the current computational study, the Peptidyl Arginine Deiminase (PADI4) receptor was taken as the target protein on the basis of different literature studies which suggested that PADI4 is involved in the citrullination of arginine residues of proteins that are directly associated with immunopathogenesis of RA (Qamar T, et al., 2021). It can even cause hyper-citrullination of histone proteins that result in inflammation of synovial membrane. Its activity is calcium dependent and occurs at gene level. PADI4 receptor is usually expressed at the site of inflammation and synovial tissues of affected individuals (Goddard TD, et al., 2005). The 3D structure of PADI4 was obtained from PDB.
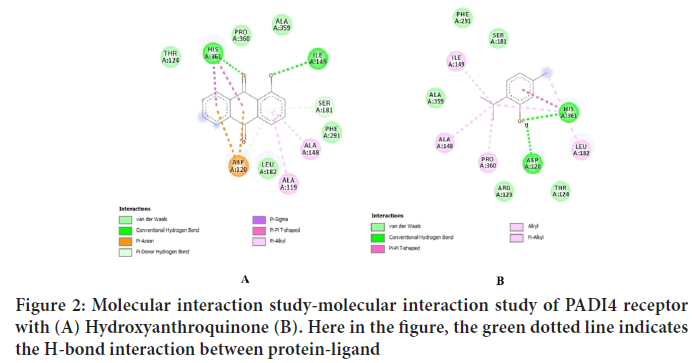
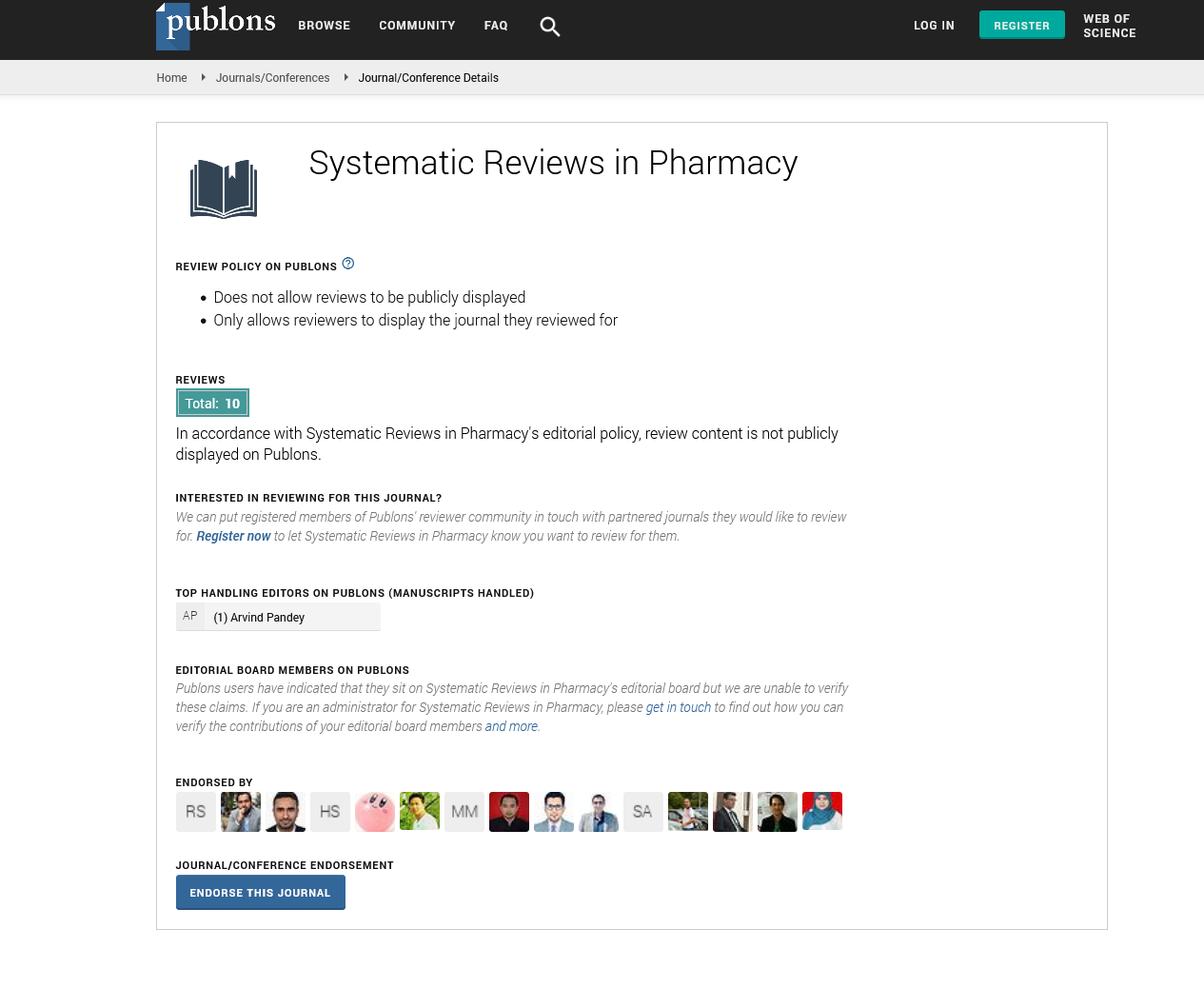
Figure 2: Molecular interaction study-molecular interaction study of PADI4 receptor with (A) Hydroxyanthroquinone (B). Here in the figure, the green dotted line indicates the H-bond interaction between protein-ligand
Figure 3: Molecular interaction study-molecular interaction study of PADI4 receptor with (A) Hydroxyanthroquinone (B) Thymol (C) Vallinic acid (D) Menthol (E) Caffeic acid. Here in the figure, the green dotted line indicates the H-bond interaction between protein-ligand
Figure 4: Molecular interaction study-molecular interaction study of PADI4 receptor with caffeic acid. Here in the figure, the green dotted line indicates the H-bond interaction between protein-ligand
Rigid docking study was performed with filtered natural and synthetic compounds with PADI4 receptor. By considering the lowest binding energy, H-bond and pi-pi interaction and docking confirmation, the three compounds namely hydroxyanthroquinone (-7.40 kcal/mol), thymol(-5.97 kcal/mol), and vallinic acid (-5.90 kcal/mol) were screened out that shows very strong binding affinity with the active site of PADI4 receptor (Huang X, et al., 2019). These phytochemicals will be cost effective and can be taken as a prophylactic approach against this malady as it prevents the disease progression by inhibiting the functions of our PADI4 receptor such that it doesn't citrullinate the histone and the protein residues for causing pathogenesis.
Conclusion
The bioactivity and drug-likeness properties of compounds show their better pharmacological property and safe to use. Hydroxyanthraquinones derived from Justicia gendarussa, have anti-inflammatory properties and are involved in suppression of pro-inflammatory cytokines secretion such as TNF-, IL-1, IL-6. They inhibit the secretion of destructive metalloproteinases that are involved in degradation of cartilage, synovial hyperplasia. Moreover, it downregulates the expression of transcription factor NF-κ B, which is capable of controlling the secretion of pro-inflammatory cytokines. Thus, they overall improve the histological architecture of rheumatic joints by limiting the new blood vessel formation within the synovial membrane and by inhibiting all the changes that are associated with bone and cartilage erosion.
Thymol derived from Thymus vulgaris, exhibits excellent anti-inflammatory activities by repressing the cytokines (IL-4, IL-5, IL-6, TNF-α) involved in pro-inflammatory responses (Li, et al., 2017). They do so by down regulating the level of nitric oxide via Inducible Nitric Oxide Synthase (iNOS) enzyme that is involved in inflammatory cascade. The anti-arthritic activity of thymol is based on its ability to suppress the NF-κ B signaling pathway and phosphorylation of Mitogen-Activated Protein Kinases (MAPKs). They target these two transcription factors as they are central players in pathophysiology of rheumatoid arthritis. Thus, they contribute to depletion of inflammatory responses through modulation of the signaling molecule exp ression like STAT-3, NF-κ B etc. Vallinic acid derived from Salix alba and is known for its ability of managing various inflammatory diseases. They inhibit the cascade pathway of PGE2, NO, COX-2 (pro-inflammatory genes) involved in immunopathogenesis of rheumatoid arthritis. By suppressing the expression of COX-2 genes that are involved in suppression of inflammatory mediators, they control the destruction of cartilage and synovial tissues. Thus, they are known for their excellent anti-inflammatory properties. The study therefore concludes the high potential of phytochemicals Hydroxyanthroquinone, thymol and vallinic acid against PADI4 receptor protein in rheumatoid arthritis. The molecular docking study depicts the better binding stability, and structural conformation at the site of binding of these compounds against the PADI4 receptor. Therefore, the above findings will definitely help in designing a novel potent PADI4 therapeutic inhibitor in case of rheumatoid arthritis. These finding may lead a better insight towards therapeutic and prophylactic approaches against RA.
References
- Spengler J, Lugonja B, Ytterberg A, Zubarev R, Creese A, Pearson M, et al. Release of active peptidyl arginine deiminases by neutrophils can explain production of extracellular citrullinated autoantigens in rheumatoid arthritis synovial fluid. Arthr Rheumatol. 2015; 67(12): 3135-3145.
[Crossref] [Google scholar] [Pubmed]
- Boissier MC, Semerano L, Challal S, Saidenberg Kermanac’h N, Falgarone G. Rheumatoid arthritis: From autoimmunity to synovitis and joint destruction. J Autoimmun. 2012; 39(3): 222-228.
[Crossref] [Google scholar] [Pubmed]
- Moussaieff A, Shohami E, Kashman Y, Fride E, Schmitz ML, Renner F, et al. Incensole acetate, a novel anti-inflammatory compound isolated from boswellia resin, inhibits nuclear factor-κ b activation. Mol Pharmacol. 2007; 72: 1657-1664.
[Crossref] [Google scholar] [Pubmed]
- Auger I, Charpin C, Balandraud N, Martin M, Roudier J. Autoantibodies to PAD4 and BRAF in rheumatoid arthritis. Autoimmun Rev. 2012; 11(11): 801-803.
[Crossref] [Google scholar] [Pubmed]
- Burmester GR, Pope JE. Novel treatment strategies in rheumatoid arthritis. Lancet. 2017; 389(10086): 2338-2348.
[Crossref] [Google scholar] [Pubmed]
- Kourilovitch M, Maldonado CG, Prado EO. Diagnosis and classification of rheumatoid arthritis. J Autoimmun. 2014; 48: 26-30.
[Crossref] [Google scholar] [Pubmed]
- Smolen J, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet. 2016; 388: 2023-2038.
[Crossref] [Google scholar] [Pubmed]
- Turunen S, Huhtakangas J, Nousiainen T, Valkealahti M, Melkko J, Risteli J, et al. Rheumatoid arthritis antigens homocitrulline and citrulline are generated by local myeloperoxidase and peptidyl arginine deiminases 2, 3 and 4 in rheumatoid nodule and synovial tissue. Arthritis Res Ther. 2016; 18(1): 239.
[Crossref] [Google scholar] [Pubmed]
- Baji P, Péntek M, Czirják L, Szekanecz Z, Nagy G, Gulácsi L, et al. Efficacy and safety of infliximab biosimilar compared to other biological drugs in rheumatoid arthritis: A mixed treatment comparison. Eur J Health Econ. 2014; 15(Suppl 1): S53-S64.
[Crossref] [Google scholar] [Pubmed]
- Laurent L, Anquetil F, Clavel C, Ndongo-Thiam N, Offer G, Miossec P, et al. IgM rheumatoid factor amplifies the inflammatory response of macrophages induced by the rheumatoid arthritis-specific immune complexes containing anticitrullinated protein antibodies. Ann Rheum Dis. 2015; 74(7): 1425-1431.
[Crossref] [Google scholar] [Pubmed]
- Gilbert D. Software review: Bioinformatics software resources. Brief Bioinform. 2004; 5(3): 300-304.
[Crossref] [Google scholar] [Pubmed]
- Koushik S, Joshi N, Nagaraju S, Mahmood S, Mudeenahally K, Padmavathy R, et al. PAD4: Pathophysiology, current therapeutics and future perspective in rheumatoid arthritis. Expert Opin Ther Targets.2017, 21(4): 433-447.
[Crossref] [Google scholar] [Pubmed]
- Khalid Z, Sezerman OU. Computational drug repurposing to predict approved and novel drug-disease associations. J Mol Graph Model. 2018; 85: 91-96.
[Crossref] [Google scholar] [Pubmed]
- Dudley JT, Deshpande T, Butte AJ. Exploiting drug-disease relationships for computational drug repositioning. Brief Bioinform. 2011; 12(4): 303-311.
[Crossref] [Google scholar] [Pubmed]
- Vakser IA. Protein-protein docking: From interaction to interactome. Biophys. J. 2014, 107, 1785-1793.
[Crossref] [Google scholar] [Pubmed]
- Moreira IS, Fernandes PA, Ramos MJ. Protein-protein docking dealing with the unknown. J Comput Chem. 2010; 31(2): 317-342.
[Crossref] [Google scholar] [Pubmed]
- Huang YF, Yeh HY, Soo VW. Inferring drug-disease associations from integration of chemical, genomic and phenotype data using network propagation. BMC Med Genomics. 2013; 6(3): 1-14.
[Crossref] [Google scholar] [Pubmed]
- Miao Z, Westhof E. RNA structure: Advances and assessment of 3D structure prediction. Annu Rev Biophys. 2017; 46: 483-503.
[Crossref] [Google scholar] [Pubmed]
- Release of active peptidyl arginine deiminases by neutrophils can explain production of extracellular citrullinated autoantigens in rheumatoid arthritis synovial fluid
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al. AutoDock 4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009; 30(16): 2785-2791.
[Crossref] [Google scholar] [Pubmed]
- Umeda N, Matsumoto I, Kawaguchi H, Kurashima Y, Kondo Y, Tsuboi H, et al. Prevalence of soluble peptidylarginine deiminase 4 (PAD4) and anti-PAD4 antibodies in autoimmune diseases. Clin Rheumatol. 2016; 35(5): 1181-1188.
[Crossref] [Google scholar] [Pubmed]
- Bovac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013; 34(5): 267-272.
[Crossref] [Google scholar] [Pubmed]
- Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004; 11(8): 777-783.
[Crossref] [Google scholar] [Pubmed]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2018; 44(D1): D1202-D1213.
[Crossref] [Google scholar] [Pubmed]
- Savarino L, Benetti D, Baldini N, Tarabusi C, Greco M, Aloisi R, et al. A preliminary in vitro and in vivo study of the effects of new anthraquinones on neutrophils and bone remodeling. J Biomed Mater Res. 2005; 75(2): 324-332.
[Crossref] [Google scholar] [Pubmed]
- Anderson AC. The process of structure based drug design. Chem Biol. 2003; 10: 787-797.
[Crossref] [Google scholar] [Pubmed]
- Luo H, Wang J, Li M, Luo J, Peng X, Wu FX, et al. Drug repositioning based on comprehensive similarity measures and bi-random walk algorithm. Bioinformatics. 2016; 32: 2664-2671.
[Crossref] [Google scholar] [Pubmed]
- Dong LM, Zhang M, Xu QL, Zhang Q, Luo B, Luo QW, et al. Two new thymol derivatives from the roots of Ageratina adenophora. Molecules. 2017; 22 (4); 592.
[Crossref] [Google scholar] [Pubmed]
- Liu H, Song Y, Guan J, Luo L, Zhuang Z. Inferring new indications for approved drugs via random walk on drug-disease heterogenous networks. BMC Bioinformatics. 2016; 17: 539.
[Crossref] [Google scholar] [Pubmed]
- Chen JJ, Tsai YC, Hwang TL, Wang TC. Thymol, benzofuranoid, and phenylpropanoid derivatives: Anti-inflammatory constituents from Eupatorium cannabinum. J Nat Prod. 2011; 74(5): 1021-1027.
[Crossref] [Google scholar] [Pubmed]
- Kuo WL, Liao HR, Jih-Jung Chen. Biflavans, flavonoids, and a dihydrochalcone from the stem wood of muntingia calabura and their inhibitory activities on neutrophil proinflammatory responses. Molecules. 2014; 19(12): 2052120535.
[Crossref] [Google scholar] [Pubmed]
- Okimoto N, Futatsugi N, Fuji H, Suenaga A, Morimoto G, Yanai R, et al. High-performance drug discovery: Computational screening by combining docking and molecular dynamics simulations. PLoS Computational Biology. 2009; 5(10): e1000528.
- Qamar T, Mukherjee S. Genetic approaches for the diagnosis and treatment of rheumatoid arthritis through personalized medicine. Gene Reports. 2021; 23: 101173.
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005; 13(3): 473-482.
[Crossref] [Google scholar] [Pubmed]
- Huang X, Xi Y, Mao Z, Chu X, Zhang R, Ma X, et al. Vanillic acid attenuates cartilage degeneration by regulating the MAPK and PI3K/AKT/NF-κ B pathways. Eur J Pharmacol. 2019; 859: 172481-172490.
[Crossref] [Google scholar] [Pubmed]
Author Info
Tooba Qamar, Prekshi Garg and Prachi Srivastava*Citation: Qamar T: A Computational Study of PADI4 Receptor for Identifying Novel Drug Target against Rheumatoid Arthritis
Received: 15-Dec-2021 Accepted: 29-Dec-2021 Published: 05-Jan-2022, DOI: 10.31858/0975-8453.13.1.17-23
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3