Research Article - (2024) Volume 15, Issue 8
Abstract
Bladder cancer is a type of cancer that arises in the bladder’s epithelial lining. It is distinguished by the bladder’s cells growing beyond control. Esculin is a naturally occurring compound found in medicinal plants that has been shown to have therapeutic effects on a number of diseases. Its possible therapeutic effect on bladder cancer is still unidentified although. This study utilizes molecular docking and network pharmacology analysis to examine the potential of esculin for the treatment of bladder cancer. Using the Search Tool for the Retrieval of Interacting Genes (STRING) database, Protein Protein Interaction (PPI) network was created between common targets. After selecting the common hub targets, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Database for Annotation, Visualization, and Integrated Discovery (DAVID) were used to carry out Gene Ontology (GO) enrichment analysis utilizing the Shiny GO databases. Additionally, molecular docking analysis was performed to investigate the interaction between esculin and these potential therapeutic targets. The main hub targets identified through PPI network analysis were Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Tumor Necrosis Factor (TNF), Matrix Metallopeptidase 9 (MMP9) gene, Epidermal Growth Factor Receptor (EGFR), Interleukin (IL)-2, MMP3, MMP1, MMP7, MMP12 and Mitogen-Activated Protein Kinase 1 (MAPK1). KEGG pathways revealed associations with bladder cancer, IL-17 signalling pathway, prostate cancer, melanoma, TNF signalling pathway, estrogen signalling pathway, endocrine resistance, relaxin signalling pathway, central carbon metabolism in cancer, Hypoxia Inducible Factor-1 (HIF- 1) signalling pathway. Esculin’s therapeutic potential has been confirmed by the molecular docking data, which demonstrated high binding affinity between esculin and the top 10 hub targets. The present study offers significant confirmation of the basic molecular mechanism in bladder cancer treatment. According to current study, esulin shows potential as possibilities for developing of novel bladder cancer therapeutics.
Keywords
Esculin, Bladder cancer, Network pharmacology, Molecular docking
Introduction
Bladder cancer, also known as urological cancer or urinary bladder cancer, is the 10th most common cancer in the world and its incidence is steadily rising worldwide, especially in developed nations (Bray F, et al., 2018). The bladder is a hollow organ in the lower abdomen whose main purpose is to store urine received from the kidneys (via the ureter) until micturition. Specialized transitional epithelial cells lining the urinary bladder and urinary tract, known as urothelial cells, accommodate the volume of urine produced by flattening under pressure. The bladder is also lined with smooth muscle that can relax to accommodate greater volumes, as well as contract (under voluntary or reflex control) to expel urine down the urethra and out of the body (Andersson KE and Arner A, 2004). The urothelial cells lining the bladder and urinary tract are constantly exposed to environmental, potentially mutagenic agents that are filtered into the urine by the kidneys. Unsurprisingly, 90% of bladder cancer cases, especially those in the developed world, arise from these urothelial cells, mostly in the bladder but on rare occasions in the urinary tract as well. While localized forms of urothelial cancer carry an excellent prognosis, if the smooth muscle is invaded, survival rates drop significantly (Mushtaq J, et al., 2019). The most common risk factor for bladder cancer, smoking is responsible for approximately two-thirds of bladder cancers in men and one-third in women. Smokers have a fourfold increased risk of bladder cancer compared with non-smokers (Griffiths TL, 2013). Because the bladder’s function is to store urine, there is ample time for carcinogens in the urine to affect the bladder. The carcinogens associated with smoking remain in constant contact within the genitourinary system until eliminated, thus the high rate for urothelial cancers (Freedman ND, et al., 2011; Turner B and Drudge-Coates L, 2012).
Esculin, also known as aesculin, is a natural compound found in various plants, particularly in the bark of trees such as horse chestnut (Aesculus hippocastanum), as well as in some herbs like the bearberry plant. It belongs to a class of compounds known as glycosides, which are characterized by their ability to undergo hydrolysis, releasing a sugar molecule when exposed to specific enzymes or conditions (Owczarek A, et al., 2021). The chemical structure of esculin consists of a glucose molecule bound to a coumarin nucleus. This unique molecular arrangement confers several interesting properties and biological activities to esculin, making it a subject of scientific interest and investigation. Moreover, esculin exhibits significant pharmacological activities, including anti-inflammatory, antioxidant, and antimicrobial effects. These bioactivities have spurred research into its potential therapeutic applications, particularly in the treatment of inflammatory disorders, oxidative stress-related conditions, and infectious diseases (Wang SK, et al., 2022). In addition to its pharmacological properties, esculin has been studied for its role in plant defense mechanisms and as a chemical marker for taxonomic classification. Its presence in certain plant species serves as a deterrent against herbivores and pathogens, highlighting its ecological significance beyond its medicinal properties (Lay MM, et al., 2014). Esculin’s pharmacological profile makes it a promising candidate for various medicinal applications. Research suggests that it exhibits potent antioxidant effects, scavenging free radicals and protecting cells from oxidative stress (Owczarek A, et al., 2021). Moreover, its anti-inflammatory properties have been investigated for their potential in mitigating inflammation and associated conditions. Additionally, esculin has shown antimicrobial activity against a range of bacterial and fungal pathogens, making it a potential candidate for the development of novel antimicrobial agents (Anand S, et al., 2024).
Focusing on therapeutics that can target many proteins involved in a disease’s features is an important part of treating diseases. Building several networks to comprehend the relationships between gene targets, illnesses, medications, and associated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways based on systems biology and computational biology is a useful technique in network pharmacology, which is used to build such treatments (Hopkins AL, 2008; Min S, et al., 2017; Kanehisa M, 2002). Furthermore, molecular docking makes it possible to compute the binding energies between ligands and receptors, which helps to forecast appropriate binding modes (Maier JK and Labute P, 2014).
The purpose of this work was to use network pharmacology and molecular docking to explore the possible targets and mechanisms of action of esculin in the treatment of bladder cancer. It is anticipated that this method will reveal novel therapeutic targets for bladder cancer and offer a molecular foundation for the application of esculin in the illness’s management.
Materials and Methods
Obtaining esculin-related target genes
PubChem database provides the canonical chemical structure and Simplified Molecular Input Line Entry System (SMILES) format for esculin (https://pubchem.ncbi.nlm.nih.gov/) (Kim S, et al., 2023). Subsequently, esculin target genes were obtained with the help of the SwissTarget- Prediction (http://www.swisstargetprediction.ch/) and Search Tool for Interactions of Chemicals (STITCH) (http://stitch.embl.de/) databases respectively (Gfeller D, et al., 2014; Kuhn M, et al., 2007). To determine the target genes for esculin, the chosen targets were then combined and duplicate results were excluded from the analysis.
Obtaining bladder cancer-related target genes
Using the keyword “bladder cancer” as search term, bladder cancer-associated target genes were selected from the GeneCards (https://www.genecards.org/) database (Stelzer G, et al., 2016). 52.01 is the largest score value that was obtained, while 0.19 is the lowest. The potential target for bladder cancer is defined as the target with a score of not less than five. The chosen targets were combined to determine the target genes for bladder cancer and duplicate data was eliminated from the investigation.
Obtaining potential common targets
Using the online tool Venny (https://bioinfogp.cnb.csic.es/tools/venny/) 2.1, the common targets were imported into a Venn diagram to identify the overlapping potential targets for esculin and bladder cancer (Figure 1).
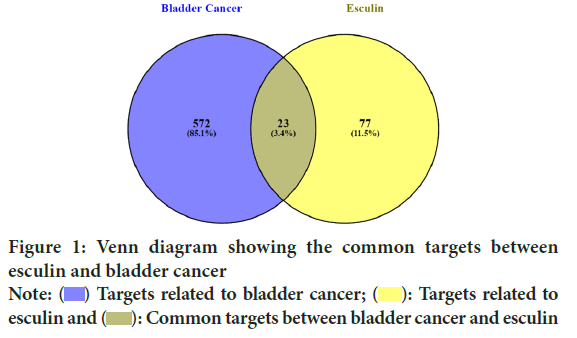
Figure 1: Venn diagram showing the common targets between
esculin and bladder cancer Note: ( ) Targets related to bladder cancer; (
) Targets related to bladder cancer; ( ): Targets related to esculin and (
): Targets related to esculin and ( ): Common targets between bladder cancer and esculin
): Common targets between bladder cancer and esculin
PPI network construction and detection of hub targets
Using STRING database, version 12.0, an interaction network was generated to explore the interaction networks of the target genes for esculin (https://string-db.org/) and the PPI network was constructed with a minimum required interaction score >0.4 (Mering CV, et al., 2003). After the collected data had been imported into Cytoscape v.3.10.2, hub genes based on the degree method and higher-degree nodes were identified using Cytoscape’s cytoHubba plugin (Saito R, et al., 2014; Shannon P, et al., 2003; Sherman BT, et al., 2022) (Figure 2).
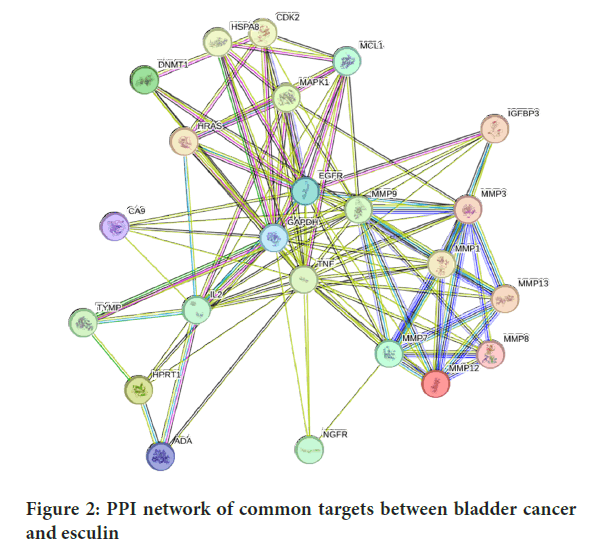
Figure 2: PPI network of common targets between bladder cancer and esculin
Pathways and GO enrichment analysis
Currently, one common method to explore genomic data-especially large- scale transcriptome data-is GO analysis. Three groups examined potential targets for GO functional enrichment, Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) (Dennis G, et al., 2003; Ge SX, et al., 2020). DAVID database (https://david.ncifcrf.gov/tools.jsp) and ShinyGO software version 0.76 (http://bioinformatics.sdstate.edu/go76/) were used to explore pathways and diseases related to the hub genes in the KEGG (https://www.genome.jp/kegg/) respectively (Kanehisa M and Goto S, 2000; Harder E, et al., 2016). Gene set enrichment results with p<0.05 was considered statistically significant.
Molecular docking
The three Dimensional (3D) structures of esculin were made with the Schrodinger Maestro v13.5 software. Using the LigPrep and Epic modules from the Schrodinger’s suite, esculin was created at physiological pH (7.0 ± 2.0). Esculin was ionized utilizing the Optimized Potentials for Liquid Simulations 4 (OPLS4) force field to produce tautomeric states after which they were converted into their 3D structures (Sastry GM, et al., 2013; Bhachoo J and Beuming T, 2017). The 3D crystal structures of target proteins were selected and acquired from the Protein Data Bank (PDB) (https:// www.rcsb.org/). Water molecules were removed from the crystal structures and hydrogen was incorporated where, it was not available. The loop gaps were closed and side chain protonation states were modified using the protein production technique. Heavier atoms were assigned hydrogens, charges and bond order. Selenomethionines were converted to methionine, once all the water was eliminated. To make the grid, the chosen protein was first broken down and then subjected to further preparation.
Subsequently, the co-crystallized enzyme ligand was surrounded by a receptor grid to reveal the binding location (Friesner RA, et al., 2006).
Non-cis/trans amide bonds were penalized during Extra Precision (XP) flexible ligand docking in Glide of Schrödinger-Maestro version 13.5 preceding High-Throughput Virtual Screening (HTVS) as a confirmation step (Bell JA, et al., 2012). The partial charge cutoff and Van der Waals scaling factor for ligand atoms were selected to be 0.15 and 0.80, respectively (Elokely KM and Doerksen RJ, 2013). Bias sampling of torsions is carried out for each of the designated functional groups and docking score was raised by applying Epic state penalties. A docking score was produced by computing the final scoring with energy-minimized poses. For every ligand, the best-docked site with the lowest docking score value was determined.
Results and Discussion
Screening of potential bladder cancer targets for esculin
We identified potential targets for esculin and bladder cancer from various databases. A database search and analysis of the SwissTargetPrediction revealed 100 possible targets linked to esculin. Following integration and deduplication of values to identify putative targets linked to esculin. Simultaneously, 3500 potential targets linked with bladder cancer were found through database querying and analysis of GeneCards and 595 prospective targets were recovered following integration, and removal of duplicate values with a score of at least 5. The possible targets for bladder cancer and esculin were then analyzed using a Venn diagram to find overlapping targets. Out of all the targets, 23 shared targets (3.4% of the total) were found.
PPI network construction and hub targets analysis
To construct the PPI network and illustrate the association between the possible targets, 23 common targets were imported into the STRING database, version 12.0. According to the findings, the PPI network has 107 edges and 23 nodes, with a clustering coefficient of 0.806 and an average node degree of 9.3. p-value of PPI enrichment was <1.0 × 10-16 and the expected number of edges was 40, which was significantly lower than the actual number of edges observed. A network diagram of the hub targets of bladder cancer and esculin was created after the PPI network and was analyzed using the cytoHubba add-on of Cytoscape software to determine the top 10 hub targets of the PPI network using the degree method (Figures 3 and 4).
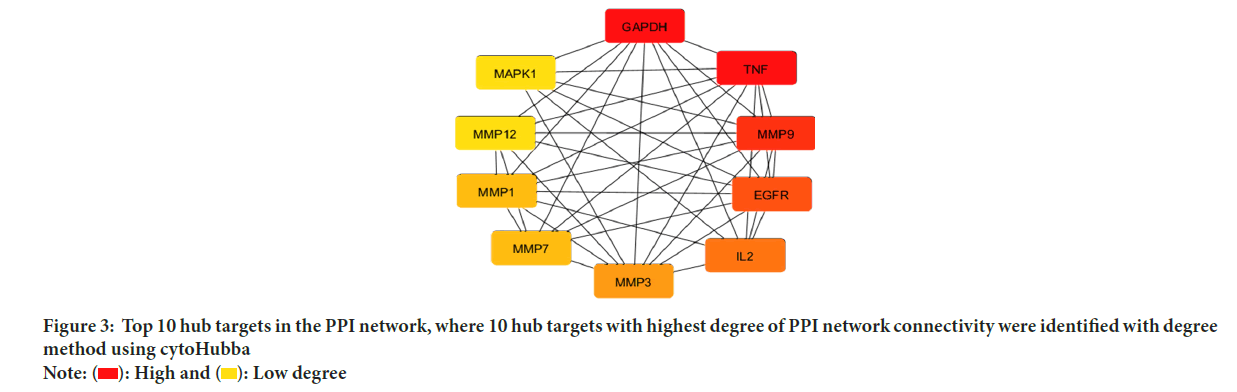
Figure 3: Top 10 hub targets in the PPI network, where 10 hub targets with highest degree of PPI network connectivity were identified with degree
method using cytoHubba Note: ( ): High and (
): High and (  ): Low degree
): Low degree
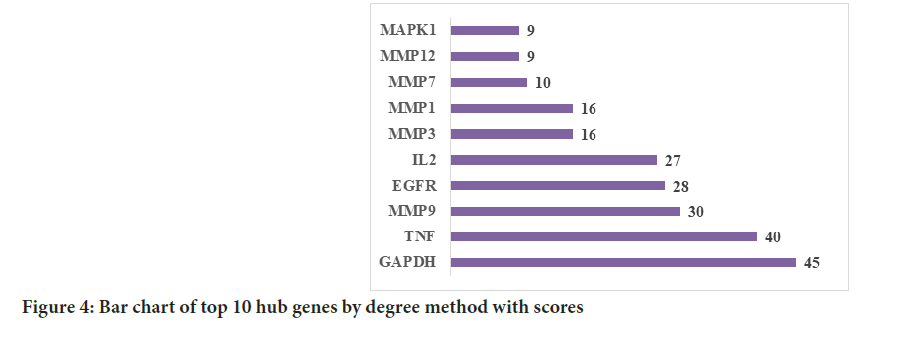
Figure 4: Bar chart of top 10 hub genes by degree method with scores
The results showed that the top 10 hub targets were GAPDH, TNF, MMP9, Epidermal Growth Factor Receptor (EGFR), IL-2, MMP3, MMP1, MMP7, MMP12 and MAPK1.
GO and KEGG pathway enrichment analysis
To understand and illustrate the molecular mechanism, GO enrichment and KEGG pathway analyses were performed using the online tools, DAVID and Shiny GO. GO enrichment analysis includes three main branches, BP, MF and CC. According to the BP, the target genes mainly involved are in cellular response to Ultraviolet-A (UV-A), cadmium ion, extracellular matrix organization, extracellular to amyloid beta, response to light stimulus, extracellular structure organization and external encapsulating structure organization, etc.
According to CC, the target genes were enriched in the Gamma Interferon Inhibitor of Translation (GAIT) complex, multi-vesicular body internal vesicle, ficolin-1-rich granule lumen, ficolin-1-rich granule, extracellular matrix, extracellular space and extracellular region (Figures 5A and 5B).
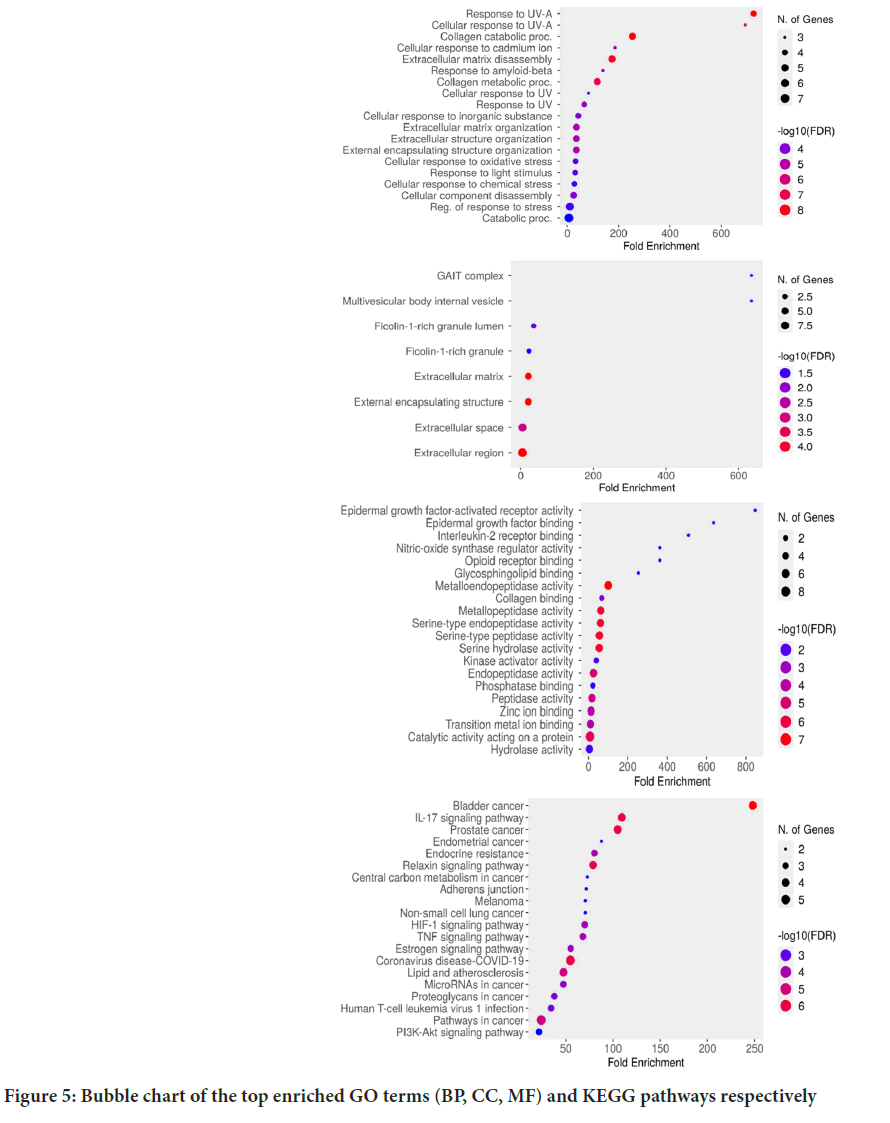
Figure 5: Bubble chart of the top enriched GO terms (BP, CC, MF) and KEGG pathways respectively
The results obtained suggest that these central targets were associated with MF, such as collagen binding, epidermal growth factor activated receptor activity, epidermal growth factor binding, IL-2 receptor binding, opioid receptor, MMP activity, serine hydrolase activity, nitric oxide synthase regulator activity, serine hydrolase activity, phosphatase binding, peptidase activity and hydrolase activity (Figures 5C and 5D).
Furthermore, the results obtained from the enrichment analysis of the KEGG pathway revealed that the predicted hub targets are related to pathways such as bladder cancer, IL-17 signalling pathway, prostate cancer, melanoma, TNF signalling pathway, estrogen signalling pathway, endocrine resistance, relaxin signalling pathway, central carbon metabolism in cancer and HIF-1 signalling pathway. Pathways with large number of common genes are clustered together in the hierarchical clustering tree, which summarizes the association among the important pathways shown in the enrichment chart (Figure 6). More significant p-values are indicated by larger dots.
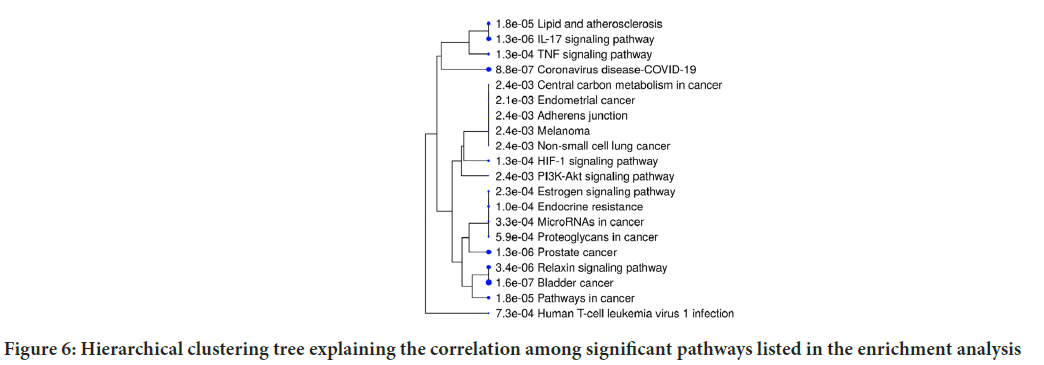
Figure 6: Hierarchical clustering tree explaining the correlation among significant pathways listed in the enrichment analysis
Molecular docking analysis
Esculin has been employed to identify the top 10 hub targets for bladder cancer; docking analysis was successful in predicting the binding energy of these targets. The target protein exhibited strong binding and high degree of matching with the top active components. The results showed that GAPDH hub target (PDB ID: 1U8F) with esculin had a score of -10.765 kcal/ mol (Figure 7), with hydrogen bridges at the following amino acids-Asparagine (ASN) Q:287, Serine (SER) O:51, Threonine (THR) Q:52, SER Q:51, ASN O:287 and Alanine (ALA) O:238. The results showed that TNF hub target (PDB ID: 1EXT) with esculin had a score of -11.050 kcal/mol (Figures 8 and 9), with hydrogen bridges at the following amino acids are Tyrosine (TYR) B:106, Arginine (ARG) B:77, Glutamic acid (GLU) A:109, ASN A:110, THR B:94, Cysteine (CYS) B:96. The results showed that MMP9 gene hub target (PDB ID: 1ITV) with esculin had a score of -9.217 kcal/mol (Figures 10 and 11), with hydrogen bridges at the following amino acids: CYS A:4, ASN A:7, ARG A:165, TYR A:187, Aspartic acid (ASP) A:195 and pi-cation bond at the ARG A:165 amino acid (Table 1).
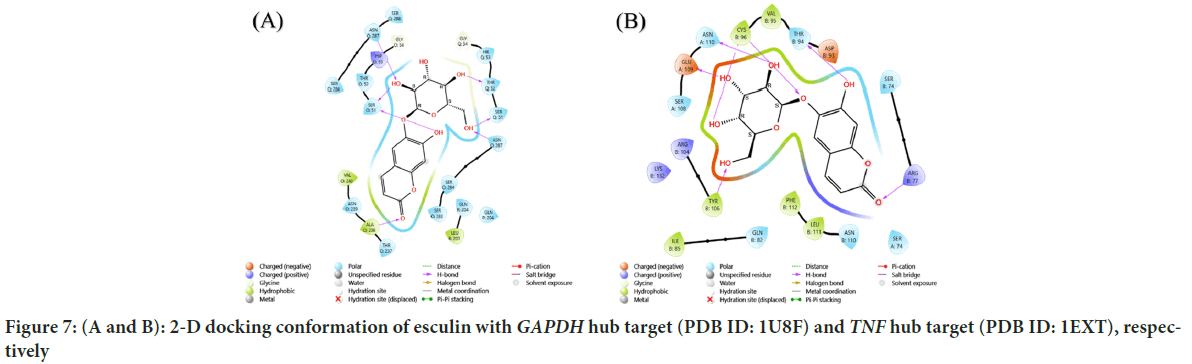
Figure 7: (A and B): 2-D docking conformation of esculin with GAPDH hub target (PDB ID: 1U8F) and TNF hub target (PDB ID: 1EXT), respectively

Figure 8: (A and B): 2-D docking conformation of esculin with MMP9 hub target (PDB ID: 1ITV) and EGFR hub target (PDB ID: 3G5Y), respectively
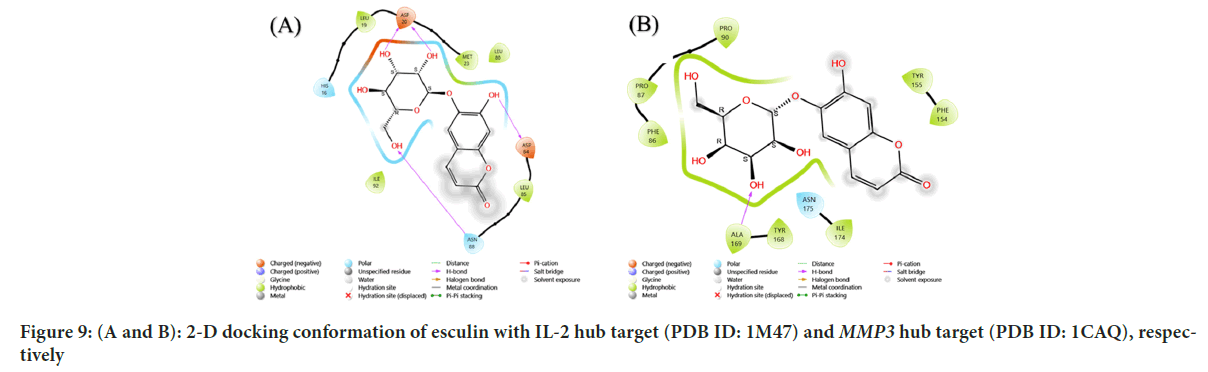
Figure 9: (A and B): 2-D docking conformation of esculin with IL-2 hub target (PDB ID: 1M47) and MMP3 hub target (PDB ID: 1CAQ), respectively
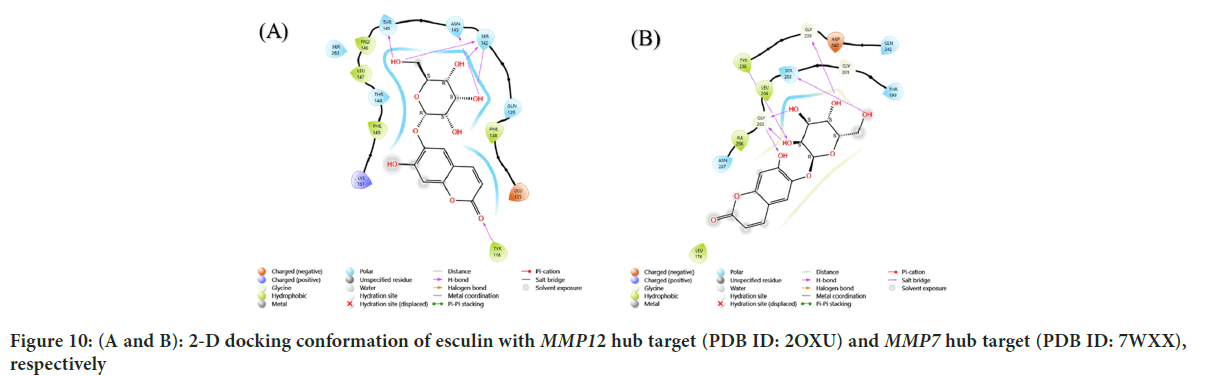
Figure 10: (A and B): 2-D docking conformation of esculin with MMP12 hub target (PDB ID: 2OXU) and MMP7 hub target (PDB ID: 7WXX), respectively
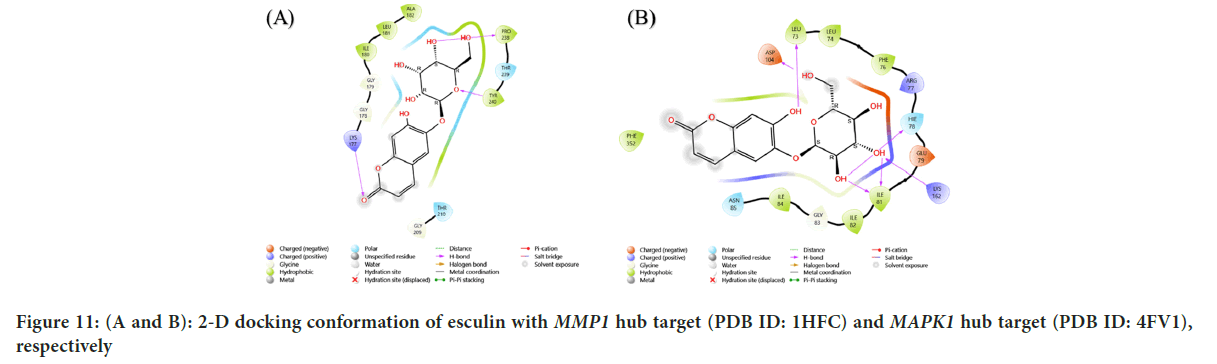
Figure 11: (A and B): 2-D docking conformation of esculin with MMP1 hub target (PDB ID: 1HFC) and MAPK1 hub target (PDB ID: 4FV1), respectively
| S. no | Hub targets | PDB ID | Docking score (kcal/mol) | Interacting residues |
|---|---|---|---|---|
| 1 | GAPDH | 1U8F | -10.765 | ASN Q:287, SER O:51, THR Q:52, SER Q:51, ASN O:287, ALA O:238 |
| 2 | TNF | 1EXT | -11.05 | TYR B:106, ARG B:77, GLU A:109, ASN A:110, THR B:94, CYS B:96 |
| 3 | MMP9 | 1ITV | -9.217 | CYS A:4, ASN A:7, ARG A:165, TYR A:187, ASP A:195 |
| 4 | EGFR | 3G5Y | -9.115 | VAL B:130, THR B:140, SER B:138, SER B:137 |
| 5 | IL-2 | 1M47 | -6.041 | THR:145, ASN:143, SER:142, TYR:116 |
| 6 | MMP3 | 1CAQ | -7.853 | GLY:205, TYR:236, SER:203, GLY:239 |
| 7 | MMP1 | 1HFC | -8.212 | THR:145, ASN:143, SER:142, TYR:116 |
| 8 | MMP7 | 7WXX | -8.331 | GLY:205, TYR:236, SER:203, GLY:239 |
| 9 | MMP12 | 2OXU | -9.183 | LYS:177, TYR:240, PRO:238 |
| 10 | MAPK1 | 4FV1 | -7.654 | ASP:104, ILE:81, LYS:162, HIE:78, LEU:73 |
Table 1: Docking score of esculin with top 10 hub targets
The results showed that EGFR hub target (PDB ID: 3G5Y) with esculin had a score of -9.115 kcal/mol, with hydrogen bridges at the following amino acids are Valine (VAL) B:130, THR B:140, SER B:138 and a metal coordination bond at the SER B:137 amino acid. The results showed that IL-2 Hub target (PDB ID: 1M47) with esculin had a score of -6.041 kcal/mol, with hydrogen bridges at the following amino acids: ASP:20, ASP:84, ASN:88. The results showed that MMP3 gene hub target (PDB ID: 1CAQ) with esculin had a score of -7.853 kcal/mol, with hydrogen bridge at the ALA:169 amino acid. The results showed that MMP12 hub target (PDB ID: 2OXU) with esculin had a score of -8.212 kcal/mol, with hydrogen bridges at the following amino acids: THR:145, ASN:143, SER:142, TYR:116. The results showed that MMP7 hub target (PDB ID: 7WXX) with esculin had a score of -8.331 kcal/mol, with hydrogen bridges at the following amino acids: GLY:205, TYR:236, SER:203, GLY:239. The results showed that MMP1 hub target (PDB ID: 1HFC) with esculin had a score of -9.183 kcal/mol, with hydrogen bridges at the following amino acids: Lysine (LYS):177, TYR:240, Proline (PRO):238. The results showed that MAPK1 hub target (PDB ID: 4FV1) with esculin had a score of -7.654 kcal/mol, with hydrogen bridges at the following amino acids, ASP:104, ILE:81, LYS:162, HIE:78, LEU:73.
Conclusion
The current study illustrates the role of esculin in the treatment of bladder cancer by combining network pharmacology and molecular docking studies. In the current study, the mechanism of known esculin and its interaction with target proteins with bladder cancer were examined using network pharmacology and molecular docking. Many signalling pathways that are crucial to esculin’s therapeutic mechanism were identified by the KEGG pathways study. These pathways include those that are involved in central carbon metabolism in cancer, TNF signaling route, estrogen signaling pathway, prostate cancer, melanoma, bladder cancer, IL-17 signaling pathway, endocrine resistance and HIF-1 signaling system.
Strong affinities for a range of hub targets and specific interactions with necessary residues suggest that these components could be utilized as building blocks for additional advancements and modifications. To completely understand the role of esculin in bladder cancer and to evaluate and enhance these findings, additional in vitro and in vivo investigations are necessary. This work lays the groundwork for future investigations into drug discovery and development, which will lead to more potent and specialized treatments. Future research into medication development and discovery will build on this work to produce more targeted and strong therapeutics.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A cancer journal for clinicians. 2018; 68(6): 394-424.
[Crossref] [Google Scholar] [Pubmed]
- Andersson KE, Arner A. Urinary bladder contraction and relaxation: Physiology and pathophysiology. Physiol Rev. 2004; 84(3): 935-86.
[Crossref] [Google Scholar] [Pubmed]
- Mushtaq J, Thurairaja R, Nair R. Bladder cancer. Surgery. 2019; 37(9): 529-537.
- Griffiths TL. Action on bladder cancer. Current perspectives in bladder cancer management. Int J Clin Pract. 2013; 67(5): 435-448.
[Crossref] [Google Scholar] [Pubmed]
- Freedman ND, Silverman DT, Hollenbeck AR, Schatzkin A, Abnet CC. Association between smoking and risk of bladder cancer among men and women. JAMA. 2011; 306(7): 737-745.
[Crossref] [Google Scholar] [Pubmed]
- Turner B, Drudge-Coates L. Bladder cancer: Risk factors, diagnosis and treatment. Cancer Nurs Pract. 2012; 11(7).
- Owczarek A, Kolodziejczyk-Czepas J, Woźniak-Serwata J, Magiera A, Kobiela N, Wąsowicz K, et al. Potential activity mechanisms of Aesculus hippocastanum bark: Antioxidant effects in chemical and biological in vitro models. Antioxidants. 2021; 10(7): 995.
[Crossref] [Google Scholar] [Pubmed]
- Wang SK, Chen TX, Wang W, Xu LL, Zhang YQ, Jin Z, et al. Aesculetin exhibited anti-inflammatory activities through inhibiting NF-кB and MAPKs pathway in vitro and in vivo. J Ethnopharmacol. 2022; 296: 115489.
[Crossref] [Google Scholar] [Pubmed]
- Lay MM, Karsani SA, Mohajer S, Malek SN. Phytochemical constituents, nutritional values, phenolics, flavonols, flavonoids, antioxidant and cytotoxicity studies on Phaleria macrocarpa (Scheff.) Boerl fruits. BMC Complement Altern Med. 2014; 14: 1-2.
[Crossref] [Google Scholar] [Pubmed]
- Owczarek A, Kołodziejczyk-Czepas J, Marczuk P, Siwek J, Wąsowicz K, Olszewska MA. Bioactivity potential of Aesculus hippocastanum L. flower: Phytochemical profile, antiradical capacity and protective effects on human plasma components under oxidative/nitrative stress in vitro. Pharmaceuticals. 2021; 14(12): 1301.
[Crossref] [Google Scholar] [Pubmed]
- Anand S, Chaudhuri A, Chopra N, Dhanorya D, Bajhaiya MK, Harsha GS, et al. A comprehensive review of therapeutical and ethnobotanical aspects, phytoconstituent and pharmacological activity of Aesculus indica. Pharmacog Res. 2024; 16(2).
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol. 2008; 4(11): 682-690.
[Crossref] [Google Scholar] [Pubmed]
- Min S, Lee B, Yoon S. Deep learning in bioinformatics. Brief Bioinform. 2017; 18(5): 851-869.
[Crossref] [Google Scholar] [Pubmed]
- Kanehisa M. The KEGG database. In ‘In silico’ simulation of biological processes: Novartis foundation symposium. 2002; 247: 91-103.
- Maier JK, Labute P. Assessment of fully automated antibody homology modeling protocols in molecular operating environment. Proteins. 2014; 82(8): 1599-1610.
[Crossref] [Google Scholar] [Pubmed]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2023 update. Nucleic Acids Res. 2023; 51(D1): D1373-D1380.
[Crossref] [Google Scholar] [Pubmed]
- Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014; 42(W1): W32-W38.
[Crossref] [Google Scholar] [Pubmed]
- Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2007; 36: D684-D688.
[Crossref] [Google Scholar] [Pubmed]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016; 54:1.30.1-1.30.33.
[Crossref] [Google Scholar] [Pubmed]
- Mering CV, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003; 31(1): 258-261.
[Crossref] [Google Scholar] [Pubmed]
- Saito R, Smoot ME, Ono K, Ruscheinski, J, Wang, PL, Lotia S, et al. A travel guide to Cytoscape plugins. BMC Syst Biol. 2014; 8: 1-7.
[Crossref] [Google Scholar] [Pubmed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11): 2498-2504.
[Crossref] [Google Scholar] [Pubmed]
- Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022; 50(W1): W216-W221.
[Crossref] [Google Scholar] [Pubmed]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003; 4: 1-1.
[Crossref] [Google Scholar] [Pubmed]
- Ge SX, Jung D, Yao R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020; 36(8): 2628-2689.
[Crossref] [Google Scholar] [Pubmed]
- Kanehisa M, Goto S. KEGG: KYOTO ENCYCLOPEDIA of GENES and GENOMES. Nucleic Acids Res. 2000; 28(1): 27-30.
[Crossref] [Google Scholar] [Pubmed]
- Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2016; 12(1): 281-296.
[Crossref] [Google Scholar] [Pubmed]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R. Ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013; 27(3): 221-234.
[Crossref] [Google Scholar] [Pubmed]
- Bhachoo J, Beuming T. Investigating protein-peptide interactions using the Schrödinger computational suite. Methods Mol Biol. 2017: 235-254.
[Crossref] [Google Scholar] [Pubmed]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006; 49(21): 6177-6196.
[Crossref] [Google Scholar] [Pubmed]
- Bell JA, Cao Y, Gunn JR, Day T, Gallicchio E, Zhou Z, et al. PrimeX and the Schrödinger computational chemistry suite of programs. International Tables for Crystallography. 2012.
- Elokely KM, Doerksen RJ. Docking challenge: Protein sampling and molecular docking performance. J Chem Inf Model. 2013; 53(8): 1934-1945.
[Crossref] [Google Scholar] [Pubmed]
Author Info
Yenumala Vamshidhar Reddy1*, Varikuppala Anand2 and Thogaru Thanish Varma32Department of Healthcare Informatics, Harrisburg University of Science and Technology, Harrisburg, Pennsylvania, United States of America
3Department of Data Analytics, Harrisburg University of Science and Technology, Harrisburg, Pennsylvania, United States of America
Citation: Reddy YV: Exploring the Therapeutic Potential of Esculin in the Treatment of Bladder Cancer
Received: 06-Aug-2024 Accepted: 22-Aug-2024 Published: 29-Aug-2024, DOI: 10.31858/0975-8453.15.8.263-270
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3