Research Article - (2024) Volume 15, Issue 3
Abstract
Exosomes, a small bilayer membrane derived from eukaryotic cells, have been identified as a useful natural delivery platform due to their suitable size, biocompatibility, structural stability, high loading capacity, and editable surface capability. Due to the difficulty of maintaining the highly pure exosome, several attempts have conducted the techniques for exosome isolation. In recent years, microstructures have found many applications in chemistry, biology, and medicine due to their high accuracy and low cost of materials. Soft lithography is a low-cost, fast, accurate, and yet widely used method of construction of Micron channels. In the present study, a soft lithography process has been performed to construct channels for exosome separation with immunoaffinity function. Both biochemical and biophysical categories tests were performed to examine the quality of extracted exosomes from different sources (serum, cell supernatant, and urine) and compared with the commercially available kit. Results showed that the current technique was capable to isolate exosomes with a high yield rate, purity, and low time consumption. All forms of the imatinib loaded exosomes exhibited the antitumor activity against KYO-1 cell line.
Keywords
Personalized medicine, Exosome, Leukemia, Targeted therapy
Introduction
Extracellular Vehicles (EVs) or exosomes have developed as one of the interesting fields due to their noteworthy role in not only biological processes but also have provided biocompatible vehicles for diagnosis and therapy (Fu S, et al., 2020). Exosomes are small size biocompatible vesicles that are able to escape from Mononuclear Phagocyte System (MPS) (Gilligan KE and Dwyer RM, 2017; He M and Zeng Y, 2016). The application of exosomes in biomedical diagnostics and therapy has highlighted the urgent need for new technologies in rapid and accurate methods of exosome separation in body fluids. Due to the biomolecular content of exosomes like proteins, nucleic acids (mRNAs, microRNAs, and DNA), numerous attempts has conducted with the diagnostic and therapeutic potential of exosomes, related to their role in cell-cell communications and drug delivery, respectively (Taylor DD and Gercel-Taylor C, 2008; Valadi H, et al., 2007; Properzi F, et al., 2013; Thind A and Wilson C, 2016; Vlassov AV, et al., 2012; Lang FM, et al., 2018). One of the main challenges is related to overcoming the fluid complexity and the lack of efficient techniques for isolation. In recent years, the field of microfluidics has made it possible to develop new methods for purifying exosomes. Microfluidics provides platforms, including micron-sized channels, for processing small amounts of fluids (microliters to picoliters). Most microfluidic devices are made with a special polymer called Poly Dimethylsiloxane (PDMS) (Xia Y and Whitesides GM, 1998). PDMS is optically transparent and biocompatible, making them a useful material in constructing biofluid devices. Microfluidic platforms can classify exosomes with a high degree of purity and sensitivity while reducing the cost, time, and volume of reagents (Talebjedi B, et al., 2021). The immunoaffinity-based microfluidic technique for exosome isolation overcomes many problems of traditional ones (such as ultracentrifugation, density gradient centrifugation, tangential flow filtration, size-exclusion chromatography) because they are adjustable, automatic, scalable, and portable (Fu S, et al., 2020; Contreras-Naranjo JC, et al., 2017; Chen J, et al., 2022; Balaj L, et al., 2015; Ghosh A, et al., 2014). Exosomes can be isolated from other components of the sample based on the same specific proteins. CD9, CD41, CD63, and CD81 as well as specific molecules like heparin, T-cell membrane protein 4 (Tim4), and heat shock protein-binding peptides are common exosome surface markers for immunoaffinity-based isolation (Cappello F, et al., 2017; Nakai W, et al., 2016). Herein, we have developed microfluidic devices for exosome isolation from different sources using CD68 conjugated magnetic beads.
Materials and Methods
In order to prepare the mask, the design of the channels with a length of 3 cm and a width of 200 µm was performed using Corel DRAW software. The desired pattern is designed to be printed using a high-resolution printer. In the next step, a layer of ultraviolet light-sensitive material with a thickness of a few micrometers is deposited on the silicon wafer surface. For SU-8-100-layer construction, 1 mm of resistant material was applied. In this regard, SU-8 is poured into silicon head for each 25 ml less than the diameter, then a spin-coating layer is applied, during which the excess material is removed from the system and the solution is allowed to evaporate. The thickness of the light-sensitive layer is determined by measuring the disk rotation speed changes. In this manner, a thin layer of resistant material is formed underneath the surface of the substrate by a two-step centrifugation procedure. In the first step, materials were stirred for the 30 s at a speed of 500 rpm, followed by 50 s at a speed of 1500 rpm. Then, with the aim of compacting the resistant material and removing the remaining materials under the layer covered with the resistant material, a three times heating treatment at 90°C was performed. Thereafter, the resistant material is selectively exposed to UV radiation (350-400 nm) for 4 minutes. The reaction created in the resistive material by the radiation changes the molecular bonds in it. After applying the radiation to form selective cross-links of the irradiated areas of the sample, the sample is baked for 2-3 minutes. The best cross-links are obtained by optimizing the irradiation and curing conditions after irradiation, after which a layer coated with a resistant material and exposed to radiation is washed in a suitable solvent for 10 to 15 minutes. After the emergence process, the substrate is washed with isopropyl alcohol and dried with nitrogen gas flow. The molding is done in vacuum conditions using the ratio of 1:10 of binders and Polydimethylsiloxane (PDMS) on the surface of micron structures and then heated up to 80°C. The polymers are coagulated for up to 30 min to form a transparent elastic solid, with micron structures on the molded surface.
COMSOL simulations
The equations governing the channels were performed using COMSOL version 5.1 Multiphysics software. The flow was simulated as a single-phase laminar flow and due to its thinness, the physical properties of distilled water at room temperature and pH 7.4 were used for the simulation. The effective and optimized parameters were set as follows: The arrangement angle of the positions along the length of the chip, the designed curved channel width, the chip height, the number of mazes, the radius of curvature of the curved channel is designed, channel width at output and input, and input flow rate.
Chip construction
Polydimethylsiloxane (Sigma, CAS no, 63148-62-9) chip mold is made of SU-8 100 patterned silicon wafer using standard soft lithography technique. SU-8 100 (Microchem Corp., Newton, MA) was stirred on a silicon wafer for 60 s at 2300 rpm. This was continued by gentle cooking at 65°C for 10 min and at 95°C for 70 min. Also, the two-step heating on wafers was performed at 65°C for 3 min and 95°C for 10 min. The wafer was then fabricated and washed using isopropanol to disable its photo resistance. To obtain the Post height of 50 μm, the wafer was then cooked at 150°C for 3 min. The microfluidic chip consists of two chambers (3500 × 2500 × 100 μm) for collecting immunomagnetic particles, two twisting channels for mixing (5000 × 500 × 100 μm), 4 inputs, and 1 output (Figure 1).
Atomic Force Microscopy (AFM)
The topography, surface roughness, phase image, friction image, magnetic properties, and thickness of the monolayer are analyzed using AFM. To investigate the changes in chip level and post and channel heights, the chip was washed with acetone and dried at 60°C.
Microfluidic-based exosome extraction
The three primary exosomal sources including serum, urine, and cell culture supernatant were considered for this part of the study. Anti-CD63 (Abcam, UK) were conjugated to Mag nanoparticles (Jiayuan Quantum Pickup Company, Wuhan, China) using conventional methods. The total binding capacity of the nanoparticles was estimated to be about 10 µg of CD63 antibody on 1 mg of nanoparticles when using 1 mg/ml of Mag-CD63. The sample containing the exosome was mixed with CD63-Mag and inserted into the microfluidic chip via input 1, and the anti-CD63 (Abcam, UK) was re-inserted through input 2. A dual syringe pump with a flow rate of 1-10 μl/min flows were mixed in the first channel, resulting in the formation of the Mag-CD63-Exo complex. Immunomagnetic particles (CD63-Mag) were retained in chamber one, by a magnetic disk. The Phosphate Buffered Saline (PBS) buffer was then introduced from input three to wash the Exo-CD63-Mag complex. It was then kept in compartment two. Immuno-conjugate exosomes were collected for examination. The isolated exosomes from serum, urine, and cellular supernatant using a chip designed r named as S-EXOChip, UE-EXOChip, and SU-EXOChip, respectively. Also, the input 4 gate is defined for drug loading, which will be discussed further.
Exosomes protein content
Bradford method was used to determine the protein content of the exosomes using 0.10, 0.08, 0.06, 0.04, 0.02 mg/ml of bovine serum albumin (BSA, Sigma, CAS no., 9048-46-8) at 595 nm (Ninfa AJ, et al., 2009).
Flow cytometry analysis
To evaluate the accuracy of the exosome extraction, flow cytometric analysis was performed against anti-CD63 antibody (Padza Padtenpajooh, cat#MM108, Iran) was performed on the extracted samples according to the manufacturer protocols. Briefly, 5 mg per test was added to the tubes containing exosomes, and then tubes were gently mixed and incubated in the dark for 60 minutes at 2°C-8°C. After that, wash the samples using 1 ml of diluted assay buffer 1×. To collect the magnetic grains, microtubes were placed in a magnetic circuit and then centrifuge at or at 3500 rpm for 10 min. Once more, 350 µl of assay buffer 1X is added to the tube and analyzed with flow cytometer (Millipore Merck Germany).
Exosome size and morphology analysis
For measuring the hydrodynamic size with DLS analysis, 200 μl of the exosomal solution of extracted cell supernatant was diluted with 420 μl of filtered PBS. The sample was then placed on ice and sonicated for 10 minutes. Finally, the sample was placed in a DLS device with 632 nm laser light beam and the results were analyzed with Zeta Sizer software. Scanning Electron Microscopy (SEM) was used to analyze the size and morphology of the exosomes. Exosomes were coated with FEI Nova 200 Nanolab Dual-beam FIB scanning electron microscope under low energy (2.0-5.0 kV) in the Electron Microscopy Analysis Laboratory (MC2) by gold spraying or carbon by thermal evaporation. Then, 200 µl of extracted cell supernatant exosomes were poured onto the slide and dried for one hour at room temperature. The sample was then observed by electron microscopy.
Western blotting
The volume of 50 μl of lysis buffer was added to both extracted samples with a 1% Holt protease inhibitor (Thermofisher scientific, LTD, USA) and incubated for 2 min at room temperature. Then, samples were kept on ice for 10 minutes and centrifuged at 13000 rpm for 15 min at 4°C. Finally, the samples were stored at -80°C for further analysis. The total volume of 20 µl of each sample was analyzed with 12% SDS Polyacrylamide Gel (SDWS-PAGE) is used to separate and determine the molecular weight of proteins with a voltage intensity of 80 mV. After separating the protein bands of the sample on SDS-PAGE gel, CD81 as a surface membrane-specific exosome protein was detected in cell lysate. In order to form an immunoblot cassette and load the gel and nitrocellulose paper on the blotting instrument (Biorad, USA). The transfer of protein bands was done for 90 min at a voltage of 100 mV. Afterward, the membrane was washed in the blocking buffer for 60 min at 37°C. Finally, the samples were stained with Anti-CD81 antibodies (1:4000) in Tris-Buffered Saline (TBS) buffer (1X), poured on PVDF membrane, and placed in a shaker incubator for 37 h at 37 °C. Then it was washed three times with TBS buffer and each time for 10 min.
Exosomal RNA extraction and miRNA expression levels
RNA extraction was performed using Norgen’s Exosome RNA Isolation Kit (Cat#58000) according to the manufacturer’s protocols. Initially, 300 μl of lysis buffer A and 37.5 μl of lysis buffer additive to each isolated exosome sample then, samples were mixed viavortexing for 10 seconds. After incubating the samples at room temperature for 10 min, 500 μl of 96% ethanol was added to the mixture and mixed well with Vertex for 10 seconds. Afterward, 500 μl of the mixture is transferred to the Mini Spin column and centrifuge at 6000 rpm for 1 min. After repeating the former step, 600 μl of washing solution A is added onto the column and centrifuge at 13000 rpm for 30 seconds, followed by centrifuge at 13000 rpm for 1 minute. Finally, the volume of 50 μl of wash solution A is added onto the column and centrifuge at 8000 rpm for 1 min and stored at -80°C for further analysis.
The total non-coding RNAs and small RNAs such as miRNAs were converted to the cDNA extraction kit (ABM good Cat#G902). The miRNA sample was prepared by mixing 2 µl of 5X poly (A) polymerase reaction buffer, 1.5 µl Adenosine Triphosphate (ATP) (10 mM), µ ml MnCl2 (25 mM), 0.5 µl Poly (A) Polymerase, Yeast (1 µg/µl), and 2.5 µl of H2O. Then, the mixture was incubated for 30 min at 37°C. Then 2 µl of miRNA Oligo (dT) adapter (10 mM) was added to the rest of the material. The mixture was incubated for 5 minutes at 65°C followed by cooling on crushed ice. Finally, 1 µl of dNTPs (10 mM), 4 µl of 5X RT buffer, 1 µl RTase (200 U/ µl) and 2 µl H2O were added to the above mixture. The cDNA synthesis was performed by incubating the samples for 15 min at 42°C and 10 min at 70°C. The microRNA content of the samples from three sources: Urine, serum, and cellular supernatant was determined using the miRCURY™ LNATM microRNA Array Hy3™/Hy5™ kit (Exiqon, Denmark) following the standard protocols. Exosome isolation using Norgen Exosome Isolation Kit (Cat#58000) from three sources: Urine, serum, and cellular supernatant. The qRT-PCR was performed for each sample in three real-time PCR System by following the manufacturer’s instructions and using different primers (Table 1). The mixture of 7 µl of Exiqon PCR Mastermix, 0.5 µl of panel primer (5 pmol/µl), 3 µl of tailed cDNA and 3 µl of enhancer, and 4 µl of DEPC Treated water were prepared. The relative expression of miRNA-155 has assessed the method with U6 as a housekeeping control.
Drug loading
The modified method of direct incubation of the drugs and CD63-Mag captured exosomes with a freeze-thaw cycle to increase is used for loading imatinib. The exosomes were incubated with the imatinib with different concentrations. 10 nM of imatinib with an injection flow rate of 1 μl/min was transferred to the input 4 of chips, at 25°C for 20 min. Then, papain and trypsin with concentrations of 0.1% and 0.02% have injected into the channel with a flow rate of 1 μl/min, respectively.
Drug release
The imatinib release was studied using a dialysis bag. 3 μg of exosome was homogenized in 3 ml of PBS buffer and then transferred to the dialysis bag and placed in a closed container with 60 ml of PBS buffer pH 7.4 and the temperature was stabilized at 37°C. The released imatinib content was calculated and at the wavelength of 242 nm using UV-visible spectroscopy. The standard curve was draw using 1, 2, 5, 10, 15, 20, and 25.30 μg/ml in PBS solution.
Cell toxicity
MTT assay is used to evaluate the toxicity of imatinib-loaded CD63-Mag captured exosomes against the KYO-1 cell line. The cellular metabolism of the cells was determined via monitoring the mitochondrial dehydrogenase enzymes activity using Methylthiazole Tetrazolium (MTT) bromide as a substrate. In this regard, 10,000 cells were pre-cultured in 96 well plates were incubated for 24 h at 37°C with 4% CO2 and 90% humidity. The exosomal samples were with different concentrations including 2, 4, 8, 10, 20, 50, 100 mg/ml were treated for 24, 48, and 72 hours. Afterward, 20 μl of MTT solution (10 mg/mL) was added to each well and incubated for 4 h in the above-mentioned condition. Finally, the supernatant was discarded and 100 μl of DMSO was added to each well, shaken for 8 minutes in a circular motion, and then the absorption of formazan at 490 nm was measured using an ELISA reader. The cell viability percentage (IC50) was examined using Graphpad Prism 6.0 software (USA).
Statistical analysis
The statistical data was analyzed by Prism 7.0 (GraphPad Software, USA). The significance of RNA quantities and qRT-PCR validation of miRNAs among exosomes was evaluated with one direct T-test. The p<0.05 was considered to be statistically significant.
Results
In the present study, we used a microfluidic system for capturing CD63 conjugated magnetic beads for highly efficient exosome separation. Polydimethylsiloxane (PDMS) chip mold is made of SU-8 50-100 patterned silicon wafer using standard soft lithography technique with immunoaffinity function (Figure 1).
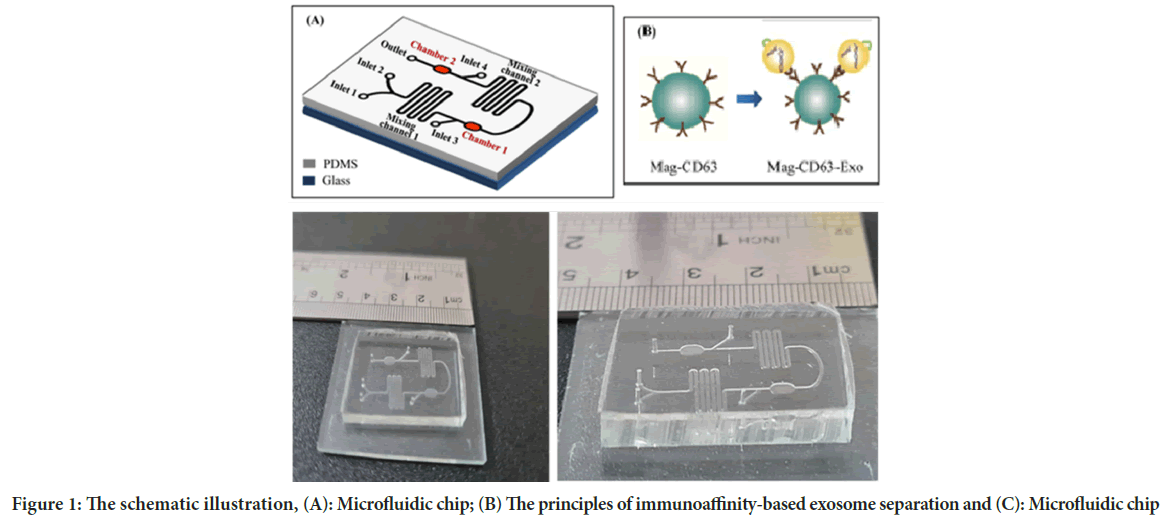
Figure 1: The schematic illustration, (A): Microfluidic chip; (B) The principles of immunoaffinity-based exosome separation and (C): Microfluidic chip
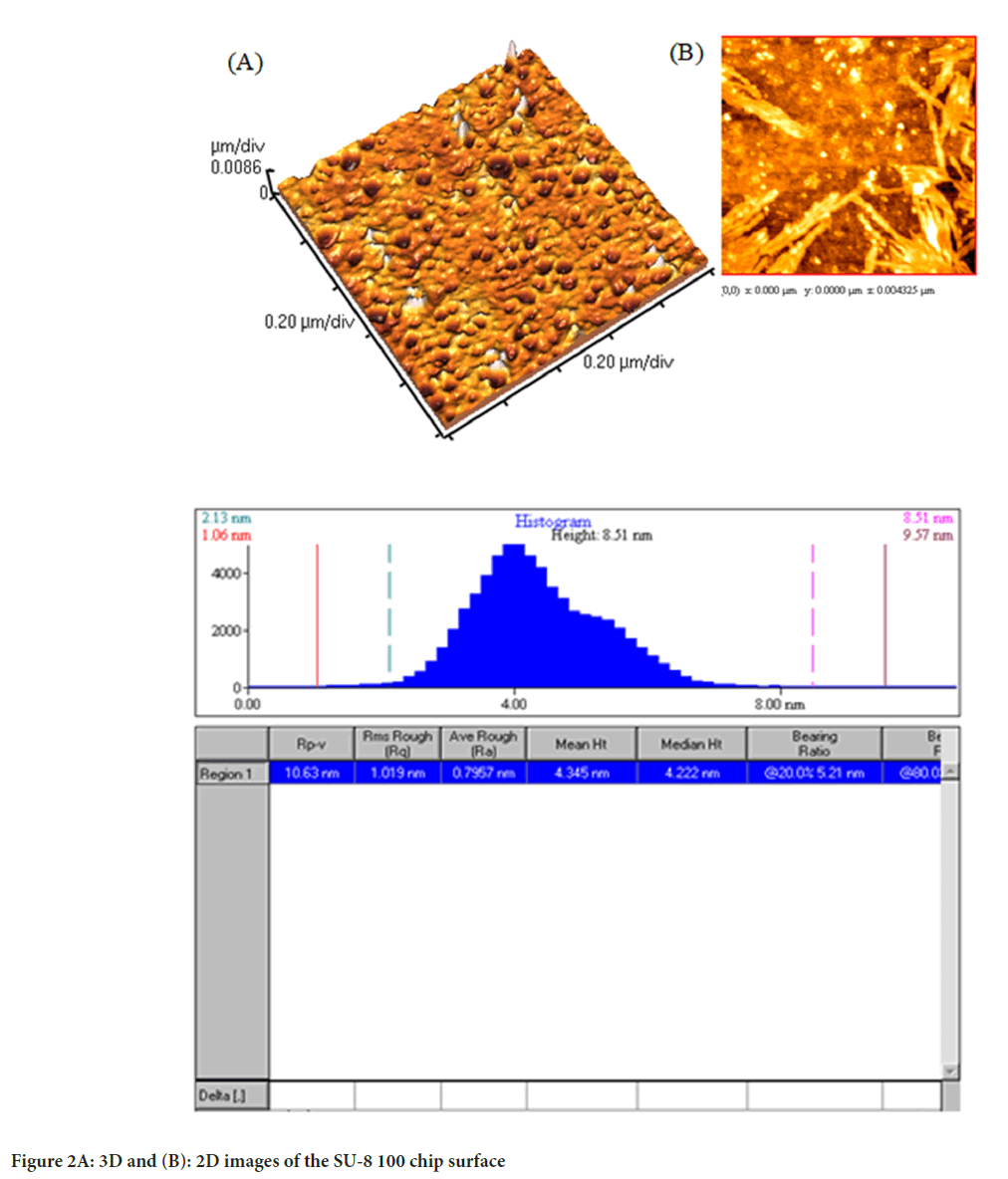
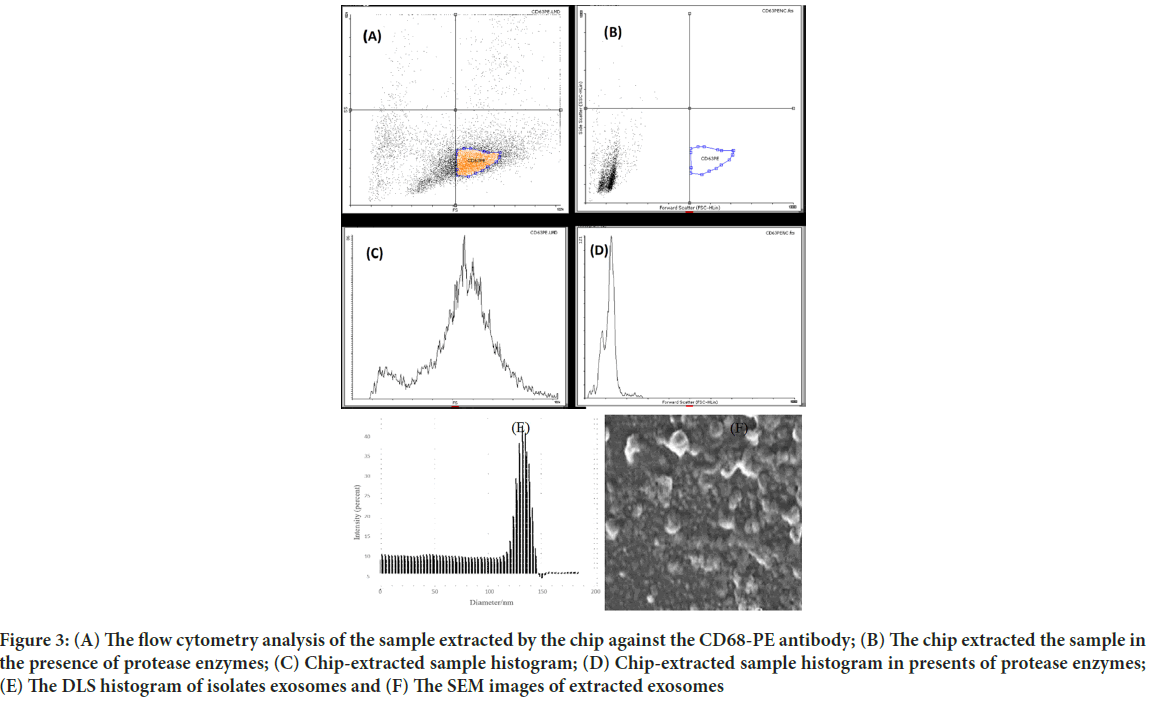
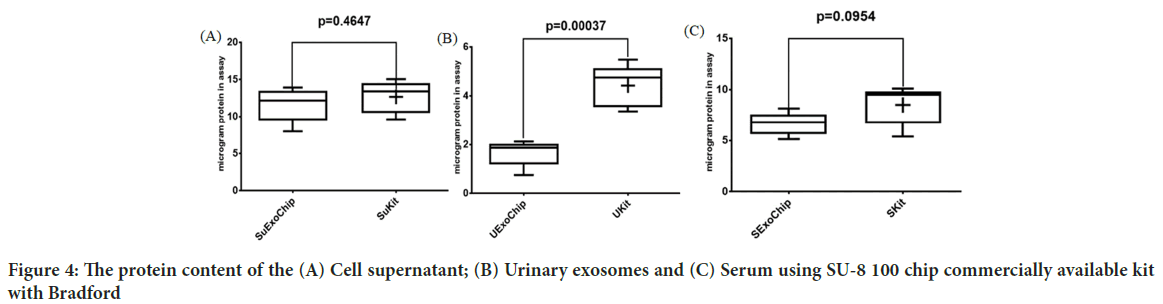
Images from AFM showed the surface of the chip. As illustrated in Figure 2, the Two-Dimensional (2D) and Three-Dimensional (3D) images of the SU-8 100 surface. After extraction of the exosomes with CD68-Mag, the physicochemical properties of the extracted exosomes were investigated using Dynamic Light Scattering (DLS), Scanning Electron Microscopy (SEM), flow cytometry, Bradford, quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR), and western blotting analysis. In this regard, exosome samples were examined by flow cytometry in the presents and absence of protease enzyme (Figures 3A-D). The hydrodynamic size of the extracted exosome from various biological sources using both commercially available kit and SU-8 100 chip was evaluated. Figure 3E, showed the hydrodynamic size distribution of extracted exosomes via DLS analysis which estimated the majority population within the mean diameter of 135 nm. The SEM analysis image is shown in Figure 3F. As it can be seen, spherical shape exosomes with a diameter between 30 and 175 nm could be detected. The comparative study of cell supernatant and urinary derived exosome protein contents using chip and kit methods with semi-quantitative method Bradford is presented in Figure 4. The protein content of serum and cellular supernatant were not significantly altered using both techniques. In the case of urinary extracted exosomes, although the commercially available protein content showed significantly higher, the proteins from the exosomes extracted by the chip are more homogeneous.
Figure 2: 3D and (B): 2D images of the SU-8 100 chip surface
Figure 3: (A) The flow cytometry analysis of the sample extracted by the chip against the CD68-PE antibody; (B) The chip extracted the sample in the presence of protease enzymes; (C) Chip-extracted sample histogram; (D) Chip-extracted sample histogram in presents of protease enzymes; (E) The DLS histogram of isolates exosomes and (F) The SEM images of extracted exosomes
Figure 4: The protein content of the (A) Cell supernatant; (B) Urinary exosomes and (C) Serum using SU-8 100 chip commercially available kit with Bradford
Investigation of the expression profile of exosome microRNAs
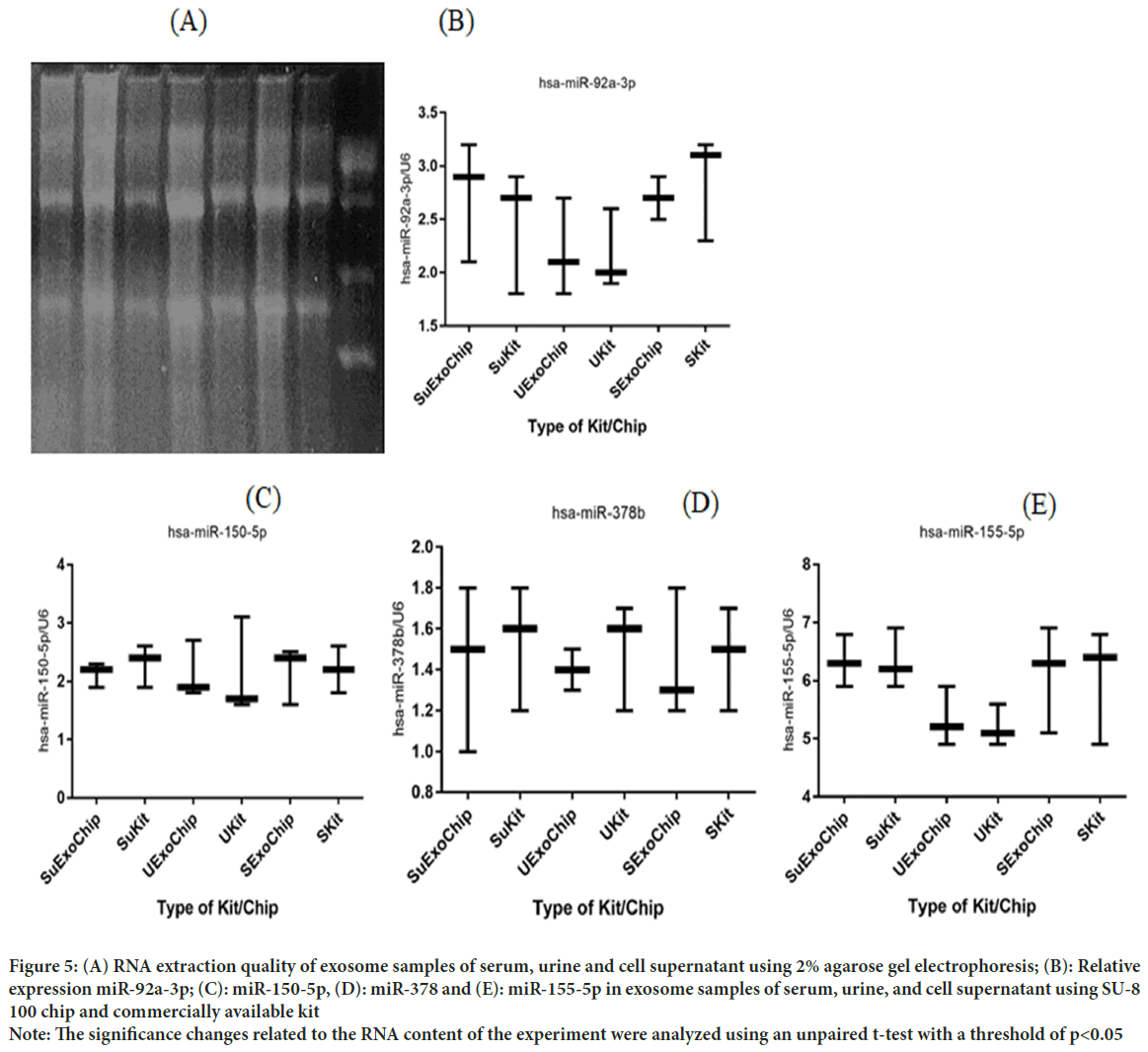
After RNA extraction was performed under RNase free conditions, agarose gel electrophoresis was performed to confirm the health of RNAs and their quality. In gel examination, the presence of ribosomal S28, S18, and S5 bands indicates no RNA degradation. In addition, the smear observed between these two bands indicates the presence of mRNA (Figure 5A-E). The diagram below shows the y-axis showing microRNA expression based on its reference gene, U6, and the horizontal axis showing the origin of the exosome samples extracted by the commercialized kit and chip designed in this study. No significant relative differences in miRNAs were observed, which indicates that the designed chip was able to isolate it without damaging the nucleic acid contents of the exosome.
Figure 5: (A) RNA extraction quality of exosome samples of serum, urine and cell supernatant using 2% agarose gel electrophoresis; (B): Relative
expression miR-92a-3p; (C): miR-150-5p, (D): miR-378 and (E): miR-155-5p in exosome samples of serum, urine, and cell supernatant using SU-8
100 chip and commercially available kit
Note: The significance changes related to the RNA content of the experiment were analyzed using an unpaired t-test with a threshold of p<0.05
Imatinib release
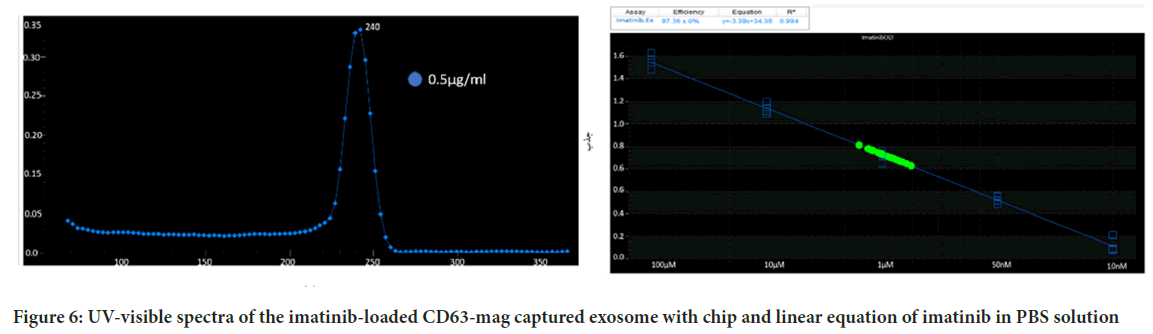
Since the preservation of the drug carrier structure such as the exosome is very effective on drug release, the release of imatinib from the extracted exosomes was investigated using both imatinib-loaded CD63-Mag captured exosome with chip. Figure 6A, showed the adsorption peak taken from imatinib solution in PBS. As can be seen, imatinib maximum absorption peak is located at 242 ± 1 nm. Calibration curve with R2=0.994 was presented to evaluate the drug release rate (Figure 6B).
Figure 6: UV-visible spectra of the imatinib-loaded CD63-mag captured exosome with chip and linear equation of imatinib in PBS solution
Cell toxicity assessments
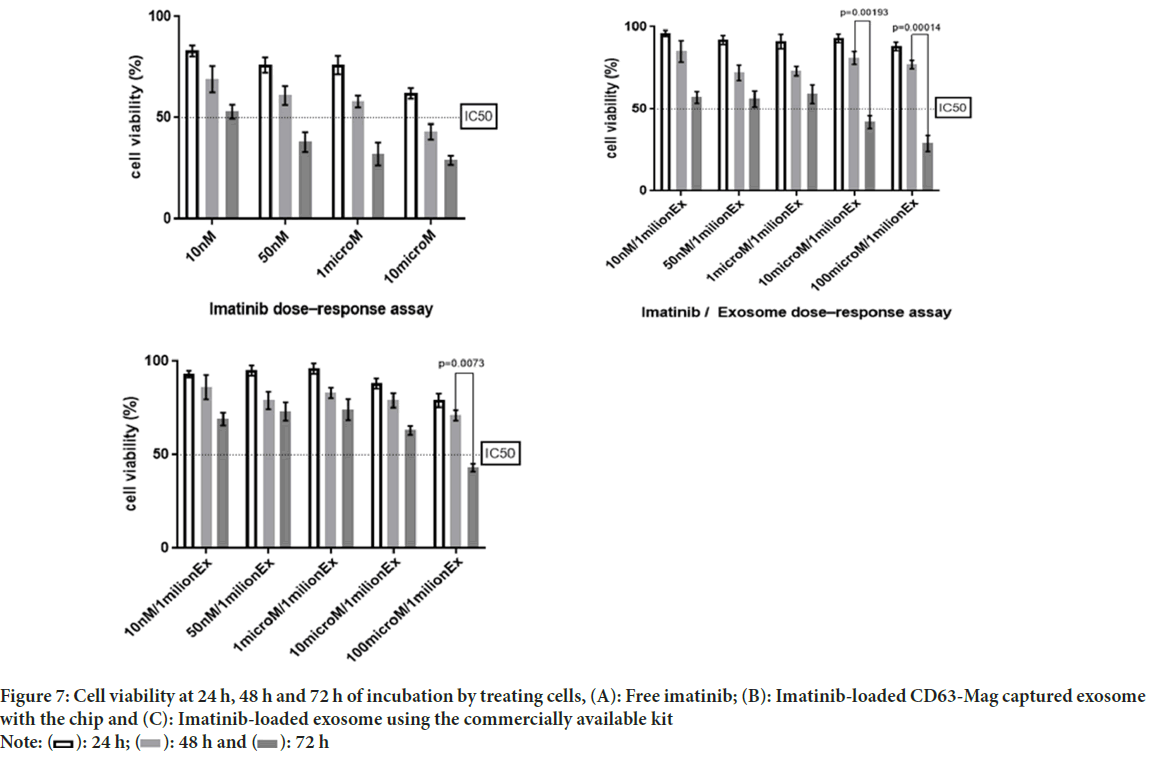
The comparison study of free imatinib, imatinib-loaded CD63-Mag captured exosome with chip and commercially available kit on the survival rate of KYO-1 cell line was done. According to Figures 7A-7C, the results indicate a dose-dependent toxicity of free imatinib, imatinib-loaded CD63-Mag captured exosome with chip and available kits on KYO-1 cells. In addition, when the cells were treated with the extracted exosome using a commercialized kit, the effective dose was changed to 100 μmol, but it is noteworthy that due to the preservation of the structure and morphology of the extracted exosome by the chip over time. The effective dose was equivalent to when imatinib was treated directly on the cells.
Figure 7: Cell viability at 24 h, 48 h and 72 h of incubation by treating cells, (A): Free imatinib; (B): Imatinib-loaded CD63-Mag captured exosome
with the chip and (C): Imatinib-loaded exosome using the commercially available kit
Discussion
In the last decade, there has been a growing interest in the role of exosomes as the fluid biopsy in the detection of cancer biomarkers. These studies have highlighted the urgent need for new technologies for the rapid and accurate separation of exosomes in body fluids with minimal sample preparation steps (Pan J, et al, 2017; Zhang X, et al., 2015; Adem B, et al., 2020; Panfoli I, 2017; Simpson RJ, et al., 2009). Exposure of exosomes to contamination and mechanical damage leads to serious issues in purity and more intact performance. The microfluidic-based exosome extraction improves the methods of many of the earlier techniques, in many aspects including isolation, molecular analysis, and detection (Chen C, et al., 2010). With the help of microfluidic technology, high-capacity exosome analyzes of up to 100 μl/min have been obtained with detection limits of about 50 exosomes/μl (He M and Zeng Y, 2016; Contreras-Naranjo JC, et al., 2017; Zhu Q, et al., 2018).
In the present study, the chip is designed with resistors that are cross-linked and tightened by placing a mask and UV light. Solvents are washed away and depressions are created with an initial layer thickness of about 100 micrometers. The CD63 antibodies for immunoaffinity-based isolation are selected as capturing molecules on magnetic beads to achieve highly efficient exosome separation. Our results showed that the device is suitable for processing different types of samples. Overall, by using and combining several methods, including engineering and design of the microfluidic system, magnetic beads, and CD63 antibody, exosome separation was used for the first time to maintain the structure of this vesicle as a drug carrier. Also, the morphology and structural features of the extracted exosomes are fully preserved, which makes it possible to load the drug and make the exosome more effective as a drug carrier.
Similar to this study has been conducted using anti-CD9 immuno-affinity magnetic beads using streptavidin-biotin conjugates on plasma samples with high separation efficiency and increased sensitivity. Also, beadbased protocols do not impose a limit on the sample size (Zarovni N, et al., 2015). In another study, anti-CD9 conjugated, anti-CD81, and antibody cocktail-conjugated magnetic nanowires magnetic beads were used for immunoaffinity isolation which provided approximately threefold greater yield than conventional methods (Lim J, et al., 2019). Also, Tim4 which binds specifically to the phosphatidylserine located on exosome cell surface, showed lower contamination than those obtained using conventional methods (Nakai W, et al., 2016).
There are plenty types of the microfluidic systems for separation. For instance, separation of particles with acoustofluidic and ultrasound methods, are mainly performed depending on particle size, density and compressibility, although it harbors the drawback of high time-consuming fabrication process depends on the particles and the fluid medium (Li P, et al., 2015). In the previous report the acoustic force microfluidic systems for exosomes isolate. Their device consisted of two consecutive modules. The first module for isolation was introduced. Although the initial compartment separated the whole blood particles, (Red Blood Cells (RBCs), White Blood Cells (WBCs), and Platelets (PLT)), exosome separation was not successful on exosomes separation (Wu M, et al., 2017). The other microfluidic platforms based on nanofiltration and centrifugation, the exodisc was introduced for exosome purification. Compared to ultracentrifugation methods, this device had an overall rapid procedure with a recovery rate of 95% with relatively high impurities due to the size-related separation (Woo HK, et al., 2017). Another report highlighted the use of ciliated microstones as an exosome trapping particle. In this system, smaller particles can pass through the microstone area without being trapped. The main weakness of the system was related to the non-specific filtration and the accumulation of large particles behind the microstone area (Villarroya-Beltri C, et al., 2013).
Particularly, immunoaffinity-based particle trapping is a highly specific way for exosome separation that can be integrated into microfluidic platforms. Several studies have conducted the use of antibody-conjugated magnetic beads for exosome separation. The CD9 conjugated magnetic beads in different sizes were tested for plasma exosome separation using an external force for magnetic beads movements with 10-15 times higher efficiencies than ultracentrifuges. They have found that incubation of antibodies conjugated magnetic beads with solutions containing exosomes provided better conditions for the interaction between vesicles and ligands on the beads. In addition, bead-based protocols do not impose a limit on the sample size (Zarovni N, et al., 2015). Also, the microfluidic device based on immunoaffinity chromatography (CD63) was used for plasma exosomes from pancreatic cancer patients. The separation relies on receptors with additional input for adding lysis buffer for direct extraction of RNA and protein from the exosomes (Chen C, et al., 2010). Another approach for designing a microfluidic device was conducted using an input channel to label the exosomes with fluorescent dye to quantify the exosome on the chip. In this study, a microfluidic device was introduced as a platform for separating exosomes on the chip and a fluorescent assay for rapid quantification of exosomes (Kanwar SS, et al., 2014). Other microfluids containing photo-lithography techniques based on micro-dimensional sites of Graphene Oxide (GO)/Polypodamine (PDA) coated with protein G were investigated for ovarian cancer exosome separation (Wu M, et al., 2017). The designed system has potential to use the exosomes for diagnostic, prognostic, and therapeutic applications. The system used asymmetric field current division technology (AF4) and two exosomal subgroups including Large exosomes (exo-L) with a diameter of 90-120 nm and Small exosomes (exo-S) with a diameter of 60–80 nm and a type distinguished nanoparticles with an approximate size of 35 nanometers, known as “exomers” (Zhang P, et al., 2016).
Conclusion
In summary, we developed a simple immunoaffinity-based microfluidic approach to for exosome separation. The designed system was capable to isolate exosome with high sensitivity, high recovery rate, and purity as well as low volume required and high-speed extraction procedure. The chip is scalable for the high-throughput isolation of exosomes with drug loading capacity for clinical uses. Hence, this platform should provide a useful tool in clinical applications for personalized medicine and as personal diagnostic devices in the future.
References
- Fu S, Wang Y, Xia X, Zheng JC. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact. 2020; 20: 100261.
- Gilligan KE, Dwyer RM. Engineering exosomes for cancer therapy. Int J Mol Sci. 2017; 18(6): 1122.
[Crossref] [Google Scholar] [Pubmed]
- He M, Zeng Y. Microfluidic exosome analysis toward liquid biopsy for cancer. J Lab Autom. 2016; 21(4): 599-608.
[Crossref] [Google Scholar] [Pubmed]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008; 110(1): 13-21.
[Crossref] [Google Scholar] [Pubmed]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9(6): 654-659.
[Crossref] [Google Scholar] [Pubmed]
- Properzi F, Logozzi M, Fais S. Exosomes: The future of biomarkers in medicine. Biomark Med. 2013; 7(5): 769-778.
[Crossref] [Google Scholar] [Pubmed]
- Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016; 5(1): 31292.
[Crossref] [Google Scholar] [Pubmed]
- Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012; 1820(7): 940-948.
[Crossref] [Google Scholar] [Pubmed]
- Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, et al. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro Oncol. 2018; 20(3): 380-390.
[Crossref] [Google Scholar] [Pubmed]
- Xia Y, Whitesides GM. Soft lithography. Angew Chem Int Ed Engl. 1998; 28(1): 153-184.
[Crossref] [Google Scholar] [Pubmed]
- Talebjedi B, Tasnim N, Hoorfar M, Mastromonaco GF, de Ferraz MMM. Exploiting microfluidics for extracellular vesicle isolation and characterization: Potential use for standardized embryo quality assessment. Front Vet Sci. 2021; 7: 620809.
[Crossref] [Google Scholar] [Pubmed]
- Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab Chip. 2017; 17(21): 3558-3577.
[Crossref] [Google Scholar] [Pubmed]
- Chen J, Li P, Zhang T, Xu Z, Huang X, Wang R, et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng Biotechnol. 2022; 9: 811971.
[Crossref] [Google Scholar] [Pubmed]
- Balaj L, Atai NA, Chen W, Mu D, Tannous BA, Breakefield XO, et al. Heparin affinity purification of extracellular vesicles. Sci Rep. 2015; 5(1): 10266.
[Crossref] [Google Scholar] [Pubmed]
- Ghosh A, Davey M, Chute IC, Griffiths SG, Lewis S, Chacko S, et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PloS One. 2014; 9(10): e110443.
[Crossref] [Google Scholar] [Pubmed]
- Cappello F, Logozzi M, Campanella C, Bavisotto CC, Marcilla A, Properzi F, et al. Exosome levels in human body fluids: A tumor marker by themselves? Eur J Pharm Sci. 2017; 96: 93-98.
[Crossref] [Google Scholar] [Pubmed]
- Nakai W, Yoshida T, Diez D, Miyatake Y, Nishibu T, Imawaka N, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016; 6(1): 33935.
[Crossref] [Google Scholar] [Pubmed]
- Ninfa AJ, Ballou DP, Benore M. Fundamental laboratory approaches for biochemistry and biotechnology. John Wiley and Sons. 2009.
- Pan J, Ding M, Xu K, Yang C, Mao LJ. Exosomes in diagnosis and therapy of prostate cancer. Oncotarget. 2017; 8(57): 97693.
[Crossref] [Google Scholar] [Pubmed]
- Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: Small particle, big player. J Hematol Oncol. 2015; 8(1): 1-3.
[Crossref] [Google Scholar] [Pubmed]
- Adem B, Vieira PF, Melo SA. Decoding the biology of exosomes in metastasis. Trends Cancer. 2020; 6(1): 20-30.
[Crossref] [Google Scholar] [Pubmed]
- Panfoli I. Cancer exosomes in urine: A promising biomarker source. Transl Cancer Res. 2017; 6(S8): S1389-S1393.
- Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009; 6(3): 267-283.
[Crossref] [Google Scholar] [Pubmed]
- Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010; 10(4): 505-511.
[Crossref] [Google Scholar] [Pubmed]
- Zhu Q, Heon M, Zhao Z, He M. Microfluidic engineering of exosomes: Editing cellular messages for precision therapeutics. Lab Chip. 2018; 18(12): 1690-1703.
[Crossref] [Google Scholar] [Pubmed]
- Zarovni N, Corrado A, Guazzi P, Zocco D, Lari E, Radano G, et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015; 87: 46-58.
[Crossref] [Google Scholar] [Pubmed]
- Lim J, Choi M, Lee H, Kim YH, Han JY, Lee ES, et al. Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J Nanobiotechnology. 2019; 17(1): 1-2.
[Crossref] [Google Scholar] [Pubmed]
- Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang PH, et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci USA. 2015; 112(16): 4970-4975.
[Crossref] [Google Scholar] [Pubmed]
- Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci USA. 2017; 114(40): 10584-10589.
[Crossref] [Google Scholar] [Pubmed]
- Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, et al. Exodisc for rapid, size-selective, and efficient isolation and analysis of nanoscale extracellular vesicles from biological samples. ACS nano. 2017; 11(2): 1360-1370.
[Crossref] [Google Scholar] [Pubmed]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013; 4(1): 2980.
[Crossref] [Google Scholar] [Pubmed]
- Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014; 14(11): 1891-1900.
[Crossref] [Google Scholar] [Pubmed]
- Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016; 16(16): 3033-3042.
[Crossref] [Google Scholar] [Pubmed]
Author Info
Amir Monfaredan1, Fakher Rahim2, Gholamreza Tavoosidana1, Mohammad Hossein Modarressi3, Alaviyehsadat Hosseininasab4, Ali-Akbar Aghajani-Afrouzi5, Mahdi Shafiee Sabet6 and Elahe Motevaseli1*2Department of Health Sciences, Cihan University-Sulaymaniyah, Sulaymaniyah, Iraq
3Department of Medical Genetics, Tehran University of Medical Sciences, Tehran, Iran
4GeneDia Life Science Company, Tehran, Iran
5Department of Business Administration, Payame Noor University, Tehran, Iran
6Neurologist, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
Citation: Monfaredan A: Imatinib Loaded Body Fluids Derived Exosomes Using Microfluidics System: An In Vitro Analysis
Received: 15-Feb-2024 Accepted: 11-Mar-2024 Published: 18-Mar-2024, DOI: 10.31858/0975-8453.15.3.110-119
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3