Review Article - (2023) Volume 14, Issue 7
Abstract
During the past 40 years, innovations in drug design and quantitative structure activity relationship have been applied to agrochemistry, pharmaceutical chemistry, toxicology, and eventually most aspects of chemistry. A Quantitative Structure Activity Relationship (QSAR) has been used for the past few decades to develop a reliable statistical model for predicting the advance activities of newly and existing chemicals by incubating their relationship and physicochemical properties. As an academic tool, QSAR is used to allow for rational prediction of biological activity and physicochemical properties and intensifying and rationalizing the mechanism of action of the series of chemicals. The use of QSAR includes mathematical methods and machine learning approaches like Support Vector Machines, Linear Regression, Partial Least Squares, and Neural Networks. Review includes the application of QSAR on drug discovery, high throughput screening, anti-HIV activity and identification of viral 3CLpro and RdRp compounds in COVID-19.
Keywords
Quantitative structure activity relationship, Descriptors, Linear regression, MLR, SVR, Neural network, High throughput screening, 3CLpro, RdRp
Introduction
The most important method for examining and utilizing the relationship between the structural characteristics of chemical compounds and biological activities by the use of computational modelling and different mathematical methods is known as Quantitative Structure Activity Relationship (QSAR) (Kwon S, et al., 2019; Golbraikh A, et al., 2012). QSAR is being applied for the past few decades in the incubation of relationship and physicochemical properties of chemical substances to acquire the reliable statistical model for the prediction of the advance activities of newly and existing chemical substances. The fundamental principle of QSAR states that variation in substance’s chemical properties is accountable in the biological properties of compounds. The physico-chemical and biological activities that correspond to pharmacokinetic characteristics including absorption, distribution, metabolism, excretion, and toxicity are referred to as structural properties. In the classical QSAR studies, inhibition constant, rate constant and other biological endpoint with atomic group and molecular properties such as lipophilicity, polarizability, electronic and steric properties (Hansch analysis) or with certain structural features (free wilson analysis) have been correlated while assimilation of ligands to respective binding sites (Verma J, et al., 2010) (Figure 1).
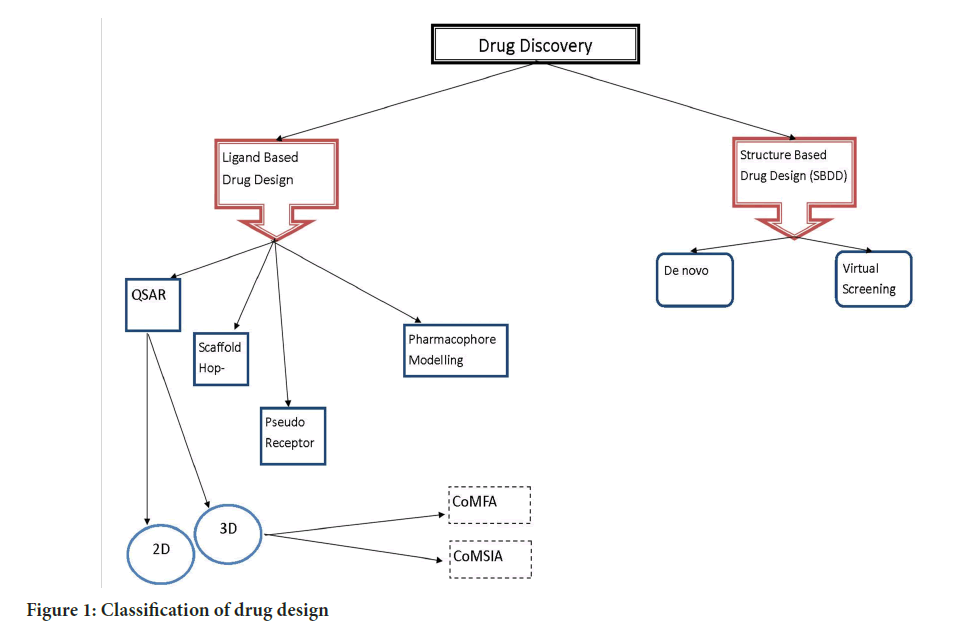
Figure 1: Classification of drug design
Literature Review
Purpose of Quantitative Structure Activity Relationship (QSAR)
QSAR is not used as the academic tool which gives allowance to post rationalisation of data. Relationships between molecular structure, chemistry, biology would be derived for a good reason. As a result of these relationships, we could create a model and with felicity judgement and expertise would be predicted.
The purpose of in silicostudies, includes the following-
• The reasonable prediction of biological activity and physicochemical characteristics.
• To clarify and strengthen the chemical series’ collective mode of action.
• The cost of manufacturing development is reduced (Example: Use in the pharmaceutical, insecticide, etc.)
• The prediction model will reduce the need for time-consuming and costly animal tests.
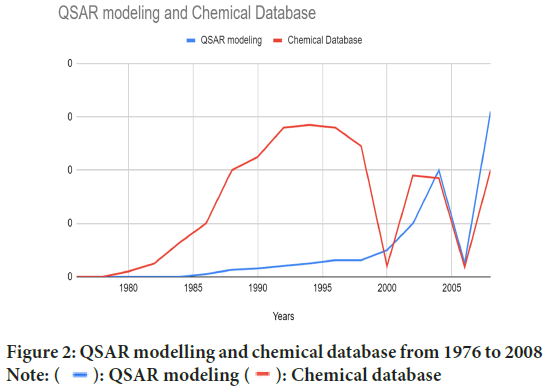
• Other initiatives to promote environmentally friendly and sustainable chemistry are less likely to be successful in increasing effectiveness or reducing waste (Borm PJ, et al., 2006; Sahdev AK, 2018) (Table 1 and Figure 2).
| Scientists name | Year of discovery | Discovery |
|---|---|---|
| Richardson BJ | 1869 | Primary alcohols' narcotic effects differ in direct proportion to their molecular weight. |
| Mills | 1884 | The melting and boiling temperatures of homologous series were predicted using QSPR models, and the findings were more accurate than one degree. |
| Richet | 1893 | The cytotoxicities of a group of alcohols, ethers, and ketones were correlated with their aqueous solubilities, and it was demonstrated that this relationship is inverse. |
| Overton and Meyer | 1897-1899 | The partition coefficients of a group of organic compounds were correlated with their anaesthetic potencies, and it was concluded that narcotic activity is dependent on molecule lipophilicity. |
| Traube J | 1904 | Found a linear relationship between narcosis and surface tension. |
| Hammet LP | 1937 | Study about chemical reactivity of substituted benzene and Hammet equation, Linear Free Energy Relationship (LFER). |
| Fergusson J | 1939 | Developed a theory connecting thermodynamics, LogP, and drug use. |
| Taft RW | 1952-1956 | Developed a method to distinguish between polar, steric, and resonance effects. |
| Hansch and Fujita | 1964 | Gave Hansch-Fujita equation for developing QSAR model |
| Free and Wilson | 1964 1970-1980 1980-1990 |
Fragments of QSAR Development of 2D QSAR Development of 3D QSAR |
Table 1: History of QSAR
Figure 2: QSAR modelling and chemical database from 1976 to 2008 Note:  : QSAR modeling
: QSAR modeling  : Chemical database
: Chemical database
40 years of innovation in drug designing, and QSAR has been applied into the practice of agrochemistry, pharmaceutical chemistry, toxicology and eventually most aspects of chemistry (Bhatia R, 2011; Hansch C, et al., 1962). In the beginning, QSAR was considered as the logical extension of physical organic chemistry. Since QSAR modelling has been applied to small series of congeneric compounds using relatively simple regression methods, it has grown and apposite into the analysis of very large datasets comprising thousands of diverse molecular structures using a wide variety of statistical and machine learning techniques (Cherkasov A, et al., 2014). Over a century ago, scientists like Crum-Brown and Fraser articulated the idea regarding the physiological action of substances which was a function of chemical composition and constitution. In 1868, Crum-Brown and Fraser expressed the term of physiological action of substance in a certain biological system (ø) and a function (f) of its chemical composition and constitution (C).
ø=f(C)
Therefore, change in chemical constitution (ΔC), would be reflected in change in biological activity (Δø) (Bhatia R, 2011; Tichý M, et al., 2008; Varnek A, 2016). Finally in 1990 virtual screening was implemented in use with QSAR.
Process in Quantitative Structure Activity Relationship (QSAR)
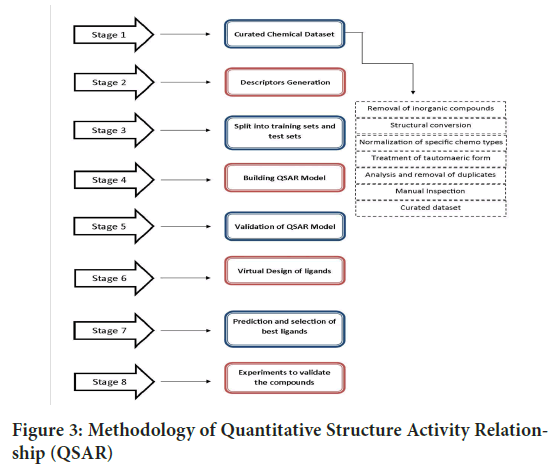
Stages involved in methodology:
1. Curation of dataset: Data curation provides a way of expressing the management of data which makes it useful for users interacting in data discovery and analysis. Data curation is an approach of creating, organizing and maintaining datasets so that whoever is looking for information can be easily accessed and used. In addition, data curation process makes the datas finable, accessible and it assists the ability to trace the information on data lineage, it classifies datas by various characteristics such as whether it is public, proprietary and protected (Guha R and Willighagen E, 2012). Steps involved in dataset curation include-
• To identify data which are required for planned analytics applications
• To map the dataset and catalogue the metadata connected with them
• To collect the datasets
• To ingest the data into a data warehouse, a data lake or other systems
• Cleans the data to fix the inconsistencies, anomalies and errors such as invalid entries, missing values, duplicate record and spelling variations.
• Model, structure and transform the data to format it for particular analytics uses.
• Create searchable indexes of the datasets to make them available for users.
• Maintain and manage the data according to ongoing analytics needs and data privacy and security requirements (Data topics, 2017)
2. Descriptor generation: The use of molecular descriptors is an effective method for approximating chemical properties in an easy-to-handle forming order to compare and select compounds that possess the characteristic that is needed by chemical, structural, pharmaceutical and biological scientists. The descriptor-based methods are not only faster than structure-based approaches, in which chemical structures or other high level structural models are compared directly against each other, but their accuracy in predicting properties is also comparable. Generation of descriptors is carried out using the following steps-
• Molecular descriptors are basically generated from molecular structures. However, various descriptors utilize diverse processing steps, still having several steps common in procedure.
• Developing molecular structure itself is the inevitable beginning step of all kinds of descriptor generation. These structures are loaded from either molecular files or databases.
• Afterwards, appropriately sized memory structures must be allocated and initialized.
• After allocating structures all the structures must be standardized.
• Another tedious process, standardization may involve aromatic ring perception, counter ion removal, and various further transformations.
• When numerous descriptors are generated simultaneously the efficiency of descriptor generation can be enhanced, as the above outlined common steps are not repeated extraneously (Chemaxon, 2023) (Figure 3). Software used in descriptor generation includes alvaDesc, Dragon, Mordred, PaDEL-descriptor etc.
Figure 3: Methodology of Quantitative Structure Activity Relationship (QSAR)
3. Split into training sets and test sets: Two-thirds of the datasets to the former dataset and one-thirds of your datasets to the latter dataset is the easiest way to split the whole datasets into training sets and test sets. Therefore, we can train the model using a training set and apply the model to the test set. In this manner, we can compare the performance of our model. On the second one observes, you should be aware of some situations while creating a model which should be considered to create an extra set called validation set. The validation set is generally required when aside from version overall performance we additionally need to select amongst fashions and evaluate which model plays better (Xu Y and Goodacre R, 2018; Myrianthous G, 2021). The software used includes panda, numpy and scikit learn.
Building of QSAR model
QSAR model basically created by using following machine learning methods-
• Multiple linear regression
• Partial least square
• Neural networks
• Support vector machine
• Gene expression programming
• Project pursuit regression etc. (Liu P and Long W, 2009)
Introduction of descriptors
Descriptors are the chemical characteristic of a molecule in numerical form. Descriptors are the outcome of a logical and mathematical process that converts the chemical information contained within a symbolic representation of a molecule into a useful number of the outcomes of a standardised experiment (Guha R and Willighagen E, 2012; Abdel-Ilah L, et al., 2017). The principal class of descriptors are presented in Table 2.
| Descriptor’s types | General information | Examples of what they measure |
|---|---|---|
| 0D | Constitutional descriptors, count descriptors | Atom types, molecular weight and bond type. |
| 1D | Structural fragments and fingerprints | Counts of atom types, counts of hydrogen bonds donor and acceptors, number of rings, number of functional groups. |
| 2D | Graph variant | Mathematical representation by graph theory and calculated values such as lipophilicity or topological polar surface area. |
| 3D | 3D-MoRSE descriptors, WHIM descriptors, GETAWAY descriptors | Geometrical descriptors or polar surface area. |
| 4D | Conformation of ligand | Non-polar and polar negative charge, hydrogen bond acceptor, hydrogen bond donor. |
Table 2: Types of descriptors
Mathematical methods used in QSAR
Quantitative structure activity connection studies are widely used in chemometrics, pharmacodynamics, pharmacokinetics, toxicology, and other fields as a common and effective research tool. Currently, mathematical strategies used to regression gear in QSAR/QSPR evaluation have been developing quickly. Therefore, by enhancing the kernel method or by combining them with other techniques, the prior strategies, such multiple Linear Regression (LR), Partial Least Square (PLS), Neural Network (NN), and Support Vector Machine (SVM), are no longer the most successful. However, further contemporary pronounced QSAR/QSPR investigations are stating some novel methods, such as Gene Expression Programming (GEP), Project Pursuit Regression (PPR), and Local Lazy Regression (LLR) (Sahdev AK, et al., 2018)
Multiple Linear Regression (MLR): One of the earliest techniques for creating QSAR/QSPR models was MLR. To this point, it has nevertheless been an often-employed maxim. The benefit of MLR is its straight- forward structure and easily understood mathematical language. MLR is susceptible to descriptors that could be associated to one another, despite being used to great effect. Therefore, it is unable to determine which associated sets are greater to the model. Recent articles that tried to improve this methodology included and proposed a few new methodologies that were entirely based on MLR. The three most significant satisfying multiples and frequently employed strategies are described in more detail below-
• Best Multiple Linear Regression (BMLR)
• Heuristic Method (HM)
• Genetic Algorithm based Multiple Linear Regression (GA-MLR)
Best Multiple Linear Regression (BMLR): To find the multi parameter regression with the best predicting potential, BMLR employs the following technique. Datasets contain all orthogonal pairs of descriptors i and j (with R2ij
Similar to MLR, BMLR is cited for having a simple and understandable mathematical statement (Katritzky AR, et al., 1996).
Heuristic Method (HM): Due of its fast calculation speed, HM, a challenging algorithm based on MLR, is widely used to build linear QSAR/QSPR equations. The advantage of HM is unquestionably based on its unique method of variable selection. Following is information about selecting descriptors-
Every descriptor is initially examined to ensure that values exist for every structure for each descriptor. When descriptions for a given value or structure are unavailable, the information is lost. Also eliminated are any descriptors whose values are the same across all of the structures in the data set. Following that, all workable one-parameter regression models are looked at, and the irrelevant descriptors are eliminated. This system then generates the pair correlation matrix of the descriptors and further decreases the descriptor pool by deleting descriptors that are strongly linked (Xia B, et al., 2009; Luan F, et al., 2006).
Genetic Algorithm based Multiple Linear Regression (GA-MLR): A novel method called as GA-MLR, which combines Genetic Algorithms (GA) and MLR, is growing in popularity in recent QSAR and QSPR studies. Using these methods, GA is completed to scan the functional region and choose the most crucial descriptors pertinent to the functions and characteristics of the substances.
The first step in GA is to generate a random collection of solutions, which is referred to as the initial population. The gene makeup of chromosomes is then used to infer a fitness feature. The Friedman LOF, which is defined as follows, is a frequently employed fitness function:
LOF=(SSE/(1-(c+dp/n))/2)2
Where,
SSE=Sum of Squares of Error
c=Number of the basis function
d=Smoothness factor
P=Number of features in model
n=Number of data points
As a result of the shortage of MLR in variable selection, GA-MLR is embedded as a well estimated parameter selection method. Similarly, to MLR, GA-MLR regression tool provides an explicit equation, as well as simple and classical regression method (Gharagheizi F, et al., 2009).
Partial Least Square (PLS): World developed the basic concept of PLS. In various field PLS is used extensively as a popular and pragmatic methodology. PLS is well-known for its use with CoMFA and CoMSIA in the field of QSAR/QSPR. The PLS method has evolved to give better performance in QSAR/QSPR analyses by combining it with other mathematical methods. There are several types of PLS-
• Genetic Partial Least Square (GPLS)
• Factor Analysis Partial Least Square (FA-PLS)
• Orthogonal Signal Correction Partial Least Square (OSC-PLS)
Genetic Partial Least Square (GPLS): Generic Function Approximation (GFA) and PLS are two methods used for QSAR calculation. The GPLS algorithm employs GFA to choose the best basis functions to incorporate into a data model, and PLS regression as the fitting method to calculate the relative contributions of the basic functions in the final model. In this way, GPLS permits the development of more substantial QSAR equations while still preventing overfitting and the elimination of the majority of variables. For the 3D-QSAR analysis tool Molecular Field Analysis (MFA), GPLS is commonly used as the regression method (Rogers D and Hopfinger AJ, 1994; Davies MN, et al., 2006).
Factor Analysis Partial Least Square (FA-PLS): Combining Factor Analysis (FA) and PLS allows for the initial descriptor selection step to be completed by FA before moving on to PLS. Factor Analysis determines how variables relate to one another. PLS regression selects important variables from the latent factors by reducing them into a few variables. When selecting the optimal number of components for PLS, a leave-one-out method is often employed (Leonard JT and Roy K, 2006).
Orthogonal Signal Correction Partial Least Square (OSC-PLS): According to Wold S, 1978 Orthogonal Signal Correction (OSC) removes systematic variations from the response matrix X that are opposite the Y-property matrix in direction. Since then, a number of OSC methods have been described in an effort to lessen model complexity through the removal of orthogonal components. Retreatment with OSC, as evidenced by many spectral analysis studies, helps traditional PLS obtain more accurate models. Unfortunately, there have been few reports of applying OSC-PLS to QSAR/QSPR studies, but it is expected that more QSAR or QSPR studies will be applied to OSC-PLS methods in the future (Yu H and MacGregor JF, 2004).
Neural Network (NN): Instead of fitting the data to the equation and reporting the coefficients derived from it, the neural network is designed to process the input information to develop a hidden relationship model. The ability of neural networks to virtually simulate nonlinear systems is a benefit. The drawbacks include the tendency to overfit the data and the challenge of identifying the most crucial descriptor in the final model. RBFNNs and GRNNs are the most frequently employed NNs in current QSAR/QSPR experiments (Xia B, et al., 2009).
• Radial Basis Function Neural Network (RBFNN)
• General Regression Neural Network (GRNN)
Support Vector Machine (SVM): It is becoming more well-known as a particular kind of system learning technique as a result of its numerous alluring features and promising empirical results. SVM initially developed for pattern reputation problems. Following that, SVM was used to regression by adding an opportunity loss feature, and the results so far are quite promising. New variations of SVM are being developed at the level of QSAR/QSPR, like-
• Least Square Support Vector Machine (LS-SVM)
• Grid Search Support Vector Machine (GS-SVM)
• Potential Support Vector Machine (P-SVM)
• Genetic Algorithms Support Vector Machine (GASVM). LS-SVM, the most commonly used one method (Cortes C and Vapnik V, 1995; Vapnik V, 1998).
Gene Expression Programming (GEP): Ferreira created gene expression programming in 1999, which developed from genetic programming and algorithms (GP). GEP makes use of the entities created by GEP (expression trees) are the expression of a genome, but they are shown in a similar way to GP’s diagrams. Compared to cell gene progression, GEP is much simpler. Chromosomes and expression trees are two specific components that are included. The process of recording gene codes and their translation is fairly straightforward along with A one-to-one relationship exists between the chromosome symbols and the features or terminals they represent. The GEP rules govern the spatial employer of capabilities and terminals within ETs, as well as the type of interaction between sub-ETs. As a result, the language of genes and ETs represents the language of GEP (Luan F, et al., 2008).
• The GEP chromosomes,
• Expression Trees (ETs)
• The mapping mechanism
Project Pursuit Regression (PPR): PPR, created by Friedman and Stuetzle, is an effective tool for looking for intriguing linear projections from high-dimensional data into lower-dimensional space. As a result, it can escape the dimensionality curse. PPR, developed by Friedman and Stuetzle, circumvented many of the issues that other nonparametric regression techniques already in use had. It does not divide the predictor space into two parts, allowing for more complicated models as needed. Additionally, because linear combinations of the predictors are represented with general smooth functions, interactions of predictor variables are immediately taken into account (Friedman JH, et al., 1984).
Local Lazy Regression (LLR): Maximum QSAR/QSPR models frequently represent the global developments in structure, activity, and property that are found across a full dataset. There are frequently subsets of molecules that exhibit a certain set of characteristics related to their activity or attribute. One may say that such a primary feature represents a close-by structure activity/property relationship. Such neighborhood relationships might not be understood by conventional models. Instead of looking at the entire dataset, LLR is a powerful technique that derives a prediction by spatially interpolating the nearby samples of the query that can be deemed relevant based on a distance degree. In light of the straightforward tenet of this approach, which is the uncomplicated presumption that comparable compounds have comparable activity or properties, it may be said that the activity or attributes of molecules will change in tandem with changes in the chemical structure. “Lazy” calculates the cost of an unknown multivariate characteristic for one or more question factors based on the notion of a collection of likely noisy samples of the feature itself.
Each sample consists of a pair of inputs and outputs, where the input is a vector and the output is an integer. The estimation of the input is obtained for each question point by fusing various nearby fashions. Local models are zeroth, first, and second-degree polynomials that match a fixed sample in the vicinity of the query point. These polynomials are taken into account for combination through lazy. According to either the “Manhattan” or the “Euclidean” distance, the neighbours are chosen. (Guha R, et al., 2006).
Validation of QSAR model
Validation process objectives to offer a version which is statistically reliable with decide on descriptors resulting from the purpose effect and no longer handiest of natural numerical received viachance but non-statistical validation which is include verification of the version in terms of the regarded mechanism of movement or other chemical information are important. It is not suited to depend upon facts most effective in validation technique. Without a doubt, that is in some way a hard procedure for cases in which no mechanism of action is known or in which records sets are small. Validation methods are wished and established predictiveness of a model. There are basically 2 types of validation methods namely, internal and external.
Internal validation depends on training datasets like Q2 (squared correlation coefficient), R2 (coefficient of determination or the coefficient of multiple determination for multiple regression), Chi-square (χ2) and Root Mean Square Error (RMSE). It shows a disadvantage as the lack of pre dictability of the model when it is applied to new datasets. While, external validation depends on testing datasets, which is considered as the best validation method (Tichý M and Rucki M, 2009; Wold S, 1978).
Application of QSAR
In drug discovery and design: Today, structure-activity studies plays more important role in drug design and development. There are many diseases for which there is no cure, so new techniques and methods are needed. Drug discovery and design requires not only discovery or design, but also drug synthesis, delivery methods, and safety assessment. The extraction of natural compounds such as morphine and cocaine are too safe for doctors to use. For these, search of less toxic, structure-based developments of known pharmacologically active compounds is necessary, which are called as tracks. SAR stands for Structure Activity Relationship while QSAR stands for Quantitative Structure Activity Relationship. SAR deals with the relationship between structure and biological activity while QSAR determines the relationship of magnitude of different structural properties with biological activity. Compounds that are structurally similar to a pharmacologically active drug are often biologically active. A lead compound SAR study can be used to determine the structure of the lead compound responsible for both its beneficial biological activity, i.e., its side effects and undesirable side effects. SAR simply makes various structural changes to make the molecule beneficial. These changes can be: Size, shape and branching of the parent structure, type of substitutes and their nature, stereochemistry of the lead compound. QSAR/QSPRs are of great importance in pharmaceutical chemistry and biochemistry because they can accelerate the development of new compounds for medicinal use (Maltarollo VG, et al., 2017; Kwon S, et al., 2019).
Virtual screening approach in QSAR: Virtual screening has emerged in drug discovery as a powerful computational approach to sifting through large libraries of small molecules to find new drugs with desirable properties that can be tested with experiment. Among virtual screening methods, QSAR is the most powerful due to its fast and high throughput. QSAR models are applied to predict the biological properties of new compounds. High-Throughput Screening (HTS) technology has led to an explosion in the amount of data suitable for QSAR models. The limitations of the above step are corrected because data retention is a required first step. Data retention procedures include removal of organics, mixtures, organic compounds, etc. HTS can rapidly identify large subsets of molecules with desired activity from a large subset of compounds using automated experimental plaque assays. Here, there are several applications of QSAR-based virtual screening to discover new results and optimise results. QSAR models for malaria were built using descriptors (0D, 1D, 2D) and Support Vector Machine methods (SVM). The current process for discovering blockbuster compounds in the early stages of drug discovery is a data-driven process, based on bioactivity data obtained from HTS campaigns. Since the cost of acquiring new successful compounds in the HTS platform is quite high, the QSAR model has played an important role in prioritizing compounds for synthesis and/or biological evaluation. QSAR models can be used for both hit to lead identification and hit to lead optimization. In the latter case, a favourable balance between potency, selectivity, and pharmacokinetic and toxicological parameters can be achieved to develop a safe and efficacious new drug through multiple optimal rounds.
Initially, the records units amassed from outside assets are curated and included to cast off or accurate inconsistent records. Using those records, QSAR fashions are evolved and proven following OECD hints and great practices of modelling. Then, QSAR fashions are used to perceive chemicals expected to be lively in opposition to decide on endpoints from huge chemical libraries. As no compound synthesis or testing is required prior to computational evaluation, QSAR represents a labour-intensive, time-consuming, and cost-effective method of obtaining compounds with specific properties and desired biology. As a result, QSAR is widely practiced in industries, universities and research centres around the world, the well-known scheme of QSAR-primarily based totally VS method. In principle, VS is frequently in comparison to a funnel, wherein a huge chemical library (i.e., a 105-107 chemical structures) is decreased through QSAR fashions to a smaller range of compounds, which then can be examined experimentally (i.e., 101-103 chemical structures). However, it is essential to say that cutting-edge VS workflows contain extra filtering steps, including: (i) Units of empirical rules (e.g., Lipinski`s (Lipinski et al., 1997) rules), (ii) chemical similarity cut-offs, (iii) different QSAR-primarily based totally filters (e.g., toxicological and pharmacokinetic endpoints), and (iv) chemical feasibility and/or purchase ability. Although the experimental validation of computational hits does now no longer constitute a part of the QSAR methodology, this must be done because it is the very last essential step. After experimental validation, a Multi-Parameter Optimization (MPO) with QSAR predictions of potency, selectivity, and pharmacokinetic parameters may be conducted. These statistics can be vital all through hit-to guide and lead optimization layout of the compound collection, to discover the properties balance (potency, selectivity, and PK) associated with the impact of various ornament styles to set up a brand-new collection of goal compounds for in vivoevaluation (Neves BJ, et al., 2018; Nantasenamat C, Prachayasittikul V, 2015; Cherkasov A, et al., 2014). QSAR study on Anti-HIV 1 activity of 4-oxo-1,4-dihydroquinoline and 4-oxo-4H-pyrido(1,2-a) pyrimidine derivatives using SW-MLR, artificial neural network and filtering method includes the following-
The causative agent of Acquired Immunodeficiency Syndrome (AIDS) is the Human Immunodeficiency Virus type 1 (HIV1). Over the past 3 decades, the combination of antiretroviral drugs in HAART (highly active antiretroviral therapy) regimens has transformed the management of HIV infection from a fatal disease to a chronic one controllability. However, resistance to commercially available antiretroviral drugs is increasing at an alarming rate. Therefore, it is necessary to develop new agents with modified scaffolds that act by different mechanisms. (Hajimahdi Z, et al., 2015; Barré-Sinoussi F, et al., 1983; Palella Jr FJ, et al., 1998).
Dataset of 25 4-oxo-1,4-dihydroquinoline and 4-oxo-4H-pyrido(1,2-a) pyrimidine derivatives were selected (Gordon DE, et al., 2020). The anti-HIV1 activities of molecules such as the rate of inhibition of p24 expression (IR) in cell culture were converted to the corresponding logarithmic inhibitory rate of p24 expression (log IR).
The total set of molecules was randomly split into a training set (20 compounds) and a test set (5 compounds) to create the QSAR model and assess the model’s efficacy.
Dragon software package for calculating descriptors are constitution descriptor, topology descriptor, molecular step counter, BCUT descriptor, Galves topology charge index, analogy 2D model, charge descriptor, aromaticity index, random molecular configuration, geometry descriptor, 3DMoRSE descriptor, WHIM descriptor, GETAWAY descriptor, experimental descriptor. 842 descriptors were analysed for the first time for the existence of constant or quasi-constant variables. Second, the correlation between descriptors and with the activity of the molecules was calculated, and the aligned descriptors (i.e., correlation coefficient between descriptors greater than 0.9) were calculated. Descriptors containing a high percentage (>90%) of the same values for all 25 molecules were removed. Among the aligned descriptors, the descriptor with the strongest correlation with the activity remained and the others were removed from the data matrix. Then, the remaining descriptors were collected in an n × m(D) data matrix, where n=25 and m=243 are the number of compounds and the number of descriptors, respectively.
Artificial Neural Network (ANN) is often applied to solve this problem because of its ability to approximate functions. In this paper, they considered a Multilayer Perceptron network (MLP) as the best-known form of ANN as a predictor to generate future output values. On the other hand, it seems that the existence of a large number of descriptors as inputs to the predictor network brings about more complexity in the MLP network. To avoid this, a stepwise variable selection step is used to select the most appropriate descriptors as network input.
The available data set is a matrix of size 20 × 243 where 20 and 243 are the total number of training groups and variables, respectively. At the end of this step, the set of best computed descriptors was selected from the dataset as input to the network to form the MLP network.
MLR analysis with stepwise selection and exclusion of variables used to associate anti-HIV 1 activity with a different descriptor. SWMLR analysis resulted in the generation of a model, with four variables (closest to the ratio of five training molecules per descriptor) and good statistical parameters for the training set and with generalizability and low prediction for the prediction set.
Described by the equation-
Log inhibition rate=-6.21(± 2.53)+0.92(± 0.14) GATS6v-25.12(± 6.36) JGI5+8.10(± 2.61) ISH-41.21(± 3.20) R6p
Discussion
Statistical parameter obtained from cross-validation (Q2) on the SWMLR model is 0.84, showing the reliability of the proposed model. IR histogram of the prediction log versus the IR of the test log, obtained using the SWM- LR model, is shown as below-
Training set: -R2 value=0.926
Test set: -R2 value=0.30
However, this method gave good results with the training set, but not with the predictive set. Therefore, they used SW-MLP to create a suitable model for both the training set and the test set. The QSAR analysis was performed on a series of 4oxo1,4dihydroquinoline and 4 oxo 4H pyrido(1,2-a) pyrimidine derivatives with the usage of the MLR and artificial neural network and filtering methods. Over 842 theoretically derived descriptors were calculated for each molecule. The best set of the calculated descriptors was selected with the stepwise method. Multiple linear regression and artificial neural networks as nonlinear systems were used for QSAR modelling. Two models exhibited good statistical qualities for the training group. In parallel, SWMLP (Nonlinear System) was found to be superior to SW- MLR in predicting test sets. Based on the results of the QSAR model, they found that electronegativity, atomic weight, atomic van der Waals volume, molecular symmetry, and polarizability are important factors controlling anti-HIV 1 activity.
QSAR machine learning models and their applications
QSAR machine learning models are for identifying viral 3CLpro- and RdRp-targeting compounds as potential therapeutics for COVID-19 and related viral infections. The ongoing COVID-19 pandemic challenges the healthcare system in many countries with high morbidity and mortality. All the SARS-CoV-2 proteins, 3 chymotrypsin-like protease (3CLpro) and RNA dependent RNA polymerase (RdRp) these 2 ideal protein targets for QSAR modelling. In addition, by comparing amino acid sequences and protein structures. 3CLpro was found to be highly conserved in SARS- CoV-2 and other human coronaviruses with sequence recognition of 96% of SARS-CoV-1, 87% for MERS-CoV and 90% with human CoV (Hajimahdi Z, et al., 2015; Ivanov J, et al., 2020; Gordon DE, et al., 2020).
RdRp is the main enzyme responsible for viral genomic RNA replication in host cells. The active site amino acid residues in RdRp are highly conserved among positive-sense single-stranded RNA ((+) ssRNA) viruses, including SARS-CoV-1 and Hepatitis C Virus (HCV) (Te Velthuis AJ, 2014)
The resulting models are used to screen 1087 FDA-approved drugs, nearly 50,000 substances from the CAS COVID-19 antiviral candidate compound dataset, and a list of 113,000 pharmacologically active substances. CAS-assigned therapeutic agents or indexed therapeutic roles in papers related to SARS, MERS and COVID19 published since 2003. Several molecules predicted from these models have been confirmed by Biologic studies and published clinical trials are a positive sign of predictive models (CAS, 2023).
3CLpro and RdRp training dataset preparation
To develop training sets for predicting SARS-CoV-2-targeting agents 3CL- pro and RdRp, we reviewed bioassay data published between 2000 and 2020 in the CAS data collection. This includes information on the substance, target, activity measurement (maximum inhibitory concentration half (IC ), maximum effective concentration half (EC ), inhibition constant (Ki) and constant of dissociation (KD), source organism and experimental details. Substances selected with test data IC50 10 µm, EC50 ≤ 10 µm, Ki ≤ 10 µm and/or KD ≤ 10 µm towards the target as active substances from this bioassay data-thresholds are suggested by researchers. Substances with IC50, EC50, Ki and KD values greater than 100 µm from this bioassay data are considered inactive (Table 3).
| Compounds name | Inhibition value (IC50) |
|---|---|
| Octapeptide AVLQSGFR | 0.031 |
| GC-373 | 3.48 |
| Rupintrivir | 3.48 |
| 1H-indole-2-carboxylic acid, 5-fluoro, 1H-benzotriazole-1-yl-ester | 0.0138 |
| GC868 | 0.4 |
| (2E)-N-[2-[[3-(Dimethylamino) propyl] thio] phenyl]-3-phenyl-2-propenamide | 4.92 |
| 3-Ethyl-2-[3-[3-[(3-ethyl-2(3H)-benzothiazoylidene) methyl]-5-methyl-2-cyclohexene-1-ylidene]-1-propen-1-yl] benzothiazolium | 2 |
Table 3: Compounds name with IC50
RdRp inhibitors of SARS-CoV-1 and other coronaviruses
For the training set for the RdRp model, we collected 1212 diverse active ingredients from 51 articles published before 2020. We also collected 67 SARS-CoV-2 RdRp inhibitors from articles published in 2020. Four unique small molecules representing the structural diversity of HCV RdRp inhibitors and two unique small molecules representing SARS-CoV-2 RdRp inhibitors are presented in Table below. DataRobot v6.0.3 https:// www.datarobot.com/, has been used to train and evaluate the performance of more than 40 different machine learning algorithms. DataRobot is a commercial tool that they used to generate selected informational features from molecular descriptors (Nguyen KT, et al., 2009). The support vector classifier (Radial Kernel) is a powerful algorithm that has been actively used to model the biological activity of small molecules for various targets (Ogura K, et al., 2019). The implementation of DataRobot is based on scikit-learn. This algorithm searches for the optimal hypersurface in the multidimensional feature space separating the classes of active and inactive compounds. The DataRobot tool determined SVM with Radial Kernel as the best model for our RdRp dataset based on the highest cross-validation values of the Area Under The Curve (AUC) among the models, another is discovered. XGBoost is a decision tree based synthetic ML algorithm using a gradient boosting framework. To generate the RdRp model, the optimised DataRobot default setting was used. Many of the composite models called “mixers” in DataRobot have also shown excellent performance comparable to the most selected models.
3CLpro model
After computing the molecular descriptors, several machine learning algorithms, including Random Forest, Gradient boosting, Neural Networks, and Support Vector Machines (SVMs), were exploited to obtain robust machine learning models. The best model was obtained using the Random Forest Classification algorithm and using Crippen logP and Morgan fingerprints as molecular representatives. This pattern has achieved a ROCAUC of 0.99, as shown earlier in the chart above. This model was then also applied to the CAS COVID19 Antiviral Compounds dataset, containing 49,437 compounds with potential antiviral activity that were identified by CAS scientists. The model predicts that these 970 chemical compounds are likely to be active against the 3CLpro coronavirus. As expected, the model identified several well-known HIV1 protease inhibitors (ritonavir and lopinavir) and identified agents (RN 2243743588, 1934276502 and 2229818464) that target the 3C/3CLpro protease and inhibit enteroviruses, MERS-CoV and SARS-CoV-1 in biological assays (Fawcett T, 2006).
RdRp model
With a probability threshold of 0.50, more than 21,000 active candidates with different structural features were found when we applied the SVM model to the three data sets described above. More than 2000 unique frame IDs were found in the predicted active candidates, indicating excellent structural diversity in the screened active complex dataset.
Conclusion
As expected, the proposed model identified several inhibitors of RdRp or polymerase (dasabuvir, cytarabine, and sofosbuvir). It has also identified several inhibitors of enzymes or receptors involved in the host immune response (ruxolitinib phosphate, duvelisib, acalabrutinib and telmisartan) and in cholesterol synthesis (Sodium fluvastatin). Among the inhibitors of the host immune response, ruxolitinib phosphate and telmisartan are being studied in multiple ongoing or upcoming COVID-19 clinical trials, including NCT04362137, NCT04348071, and NCT04360551. Thus, this model can be beneficial for the prediction of the biological activities and physicochemical properties.
References
- Kwon S, Bae H, Jo J, Yoon S. Comprehensive ensemble in QSAR prediction for drug discovery. BMC bioinformatics. 2019; 20(1): 1-2.
[Crossref] [Google Scholar] [Pubmed]
- Golbraikh A, Wang XS, Zhu H, Tropsha A. Predictive QSAR modeling: Methods and applications in drug discovery and chemical risk assessment. Springer. 2012.
- Verma J, Khedkar VM, Coutinho EC. 3D-QSAR in drug design-a review. Curr Top Med Chem. 2010; 10(1): 95-115.
[Crossref] [Google Scholar] [Pubmed]
- Borm PJ, Robbins D, Haubold S, Kuhlbusch T, Fissan H, Donaldson K, et al. The potential risks of nanomaterials: A review carried out for ECETOC. Part Fibre Toxicol. 2006; 3: 1-35.
[Crossref] [Google Scholar] [Pubmed]
- Sahdev AK, Sethi B, Rawat SL, Singh A, Anand N. A review article role of QSAR: Significance and uses in molecular design. J Emerg Technol Innov Res. 2018; 5(1): 426-435.
- Bhatia R. History in the revolution of QSAR: A review. Pharmatutor. 2011.
- Hansch C, Maloney PP, Fujita T, Muir RM. Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature. 1962; 194(4824): 178-180.
- Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, et al. QSAR modeling: Where have you been? Where are you going to? J Med Chem. 2014; 57(12): 4977-5010.
[Crossref] [Google Scholar] [Pubmed]
- Tichý M, Hanzlíková I, Rucki M, Pokorná A, Uzlová R, Tumová J. Acute toxicity of binary mixtures: Alternative methods, QSAR and mechanisms. Interdiscip Toxicol. 2008; 1(1): 15.
[Crossref] [Google Scholar] [Pubmed]
- Varnek A. Quantitative Structure-Activity Relationships Quantitative Structure-Property-Relationships (QSAR and QPSR). 2016.
- Guha R, Willighagen E. A survey of quantitative descriptions of molecular structure. Curr Top Med Chem. 2012; 12(18): 1946-1956.
[Crossref] [Google Scholar] [Pubmed]
- Data curation 101: The what, why and how. Data topics. 2017.
- Chemoinformatic and bioinformatic software development company. Chemaxon. 2023.
- Xu Y, Goodacre R. On splitting training and validation set: A comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J Anal Test. 2018; 2(3): 249-262.
[Crossref] [Google Scholar] [Pubmed]
- Myrianthous G. How to split a dataset into training and testing sets with Python. Medium. 2021.
- Liu P, Long W. Current mathematical methods used in QSAR/QSPR studies. Int J Mol Sci. 2009; 10(5): 1978-1998.
[Crossref] [Google Scholar] [Pubmed]
- Abdel-Ilah L, Veljović E, Gurbeta L, Badnjević A. Applications of QSAR study in drug design. Int J Eng Res Technol. 2017; 6(06).
- Katritzky AR, Lobanov VS, Karelson M, Murugan R, Grendze MP, Toomey JE. Comprehensive descriptors for structural and statistical analysis. 1: Correlations between structure and physical properties of substituted pyridines. Rev Roum Chim. 1996; 41(11-12): 851-867.
- Xia B, Liu K, Gong Z, Zheng B, Zhang X, Fan B. Rapid toxicity prediction of organic chemicals to Chlorella vulgaris using quantitative structure-activity relationships methods. Ecotoxicol Environ Saf. 2009; 72(3): 787-794.
[Crossref] [Google Scholar] [Pubmed]
- Luan F, Ma WP, Zhang XY, Zhang HX, Liu MC, Hu ZD, et al. QSAR study of polychlorinated dibenzodioxins, dibenzofurans, and biphenyls using the heuristic method and support vector machine. QSAR Comb Sci. 2006; 25(1): 46-55.
- Gharagheizi F, Tirandazi B, Barzin R. Estimation of aniline point temperature of pure hydrocarbons: A quantitative structure-property relationship approach. Ind Eng Chem Res. 2009; 48(3): 1678-1682.
- Rogers D, Hopfinger AJ. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci. 1994; 34(4): 854-866.
- Davies MN, Hattotuwagama CK, Moss DS, Drew MG, Flower DR. Statistical deconvolution of enthalpic energetic contributions to MHC-peptide binding affinity. BMC Struct Biol. 2006; 6: 1-3.
- Leonard JT, Roy K. Comparative QSAR modeling of CCR5 receptor binding affinity of substituted 1-(3, 3-diphenylpropyl)-piperidinyl amides and ureas. Bioorg Med Chem Lett. 2006; 16(17): 4467-4474.
[Crossref] [Google Scholar] [Pubmed]
- Wold S. Cross-validatory estimation of the number of components in factor and principal components models. Technometrics. 1978; 20(4): 397-405.
- Yu H, MacGregor JF. Post processing methods (PLS-CCA): Simple alternatives to preprocessing methods (OSC-PLS). Chemometr Intell Lab Syst. 2004; 73(2): 199-205.
- Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995; 20(3): 273-297.
- Vapnik V. The support vector method of function estimation. InNonlinear modeling: Advanced black-box techniques. Springer. 1998.
- Luan F, Si HZ, Liu HT, Wen YY, Zhang XY. Prediction of atmospheric degradation data for POPs by gene expression programming. SAR QSAR Environ Res. 2008; 19(5-6): 465-479.
[Crossref] [Google Scholar] [Pubmed]
- Friedman JH, Stuetzle W, Schroeder A. Projection pursuit density estimation. J Am Stat Assoc. 1984; 79(387): 599-608.
- Guha R, Dutta D, Jurs PC, Chen T. Local lazy regression: Making use of the neighborhood to improve QSAR predictions. J Chem Inf Model. 2006; 46(4): 1836-1847.
[Crossref] [Google Scholar] [Pubmed]
- Tichý M, Rucki M. Validation of QSAR models for legislative purposes. Interdiscip Toxicol. 2009; 2(3): 184-186.
[Crossref] [Google Scholar] [Pubmed]
- Maltarollo VG, Kronenberger T, Wrenger C, Honorio KM. Current trends in quantitative structure-activity relationship validation and applications on drug discovery. Future Sci OA. 2017; 3(4): FSO214.
- Neves BJ, Braga RC, Melo-Filho CC, Moreira-Filho JT, Muratov EN, Andrade CH. QSAR-based virtual screening: Advances and applications in drug discovery. Front Pharmacol. 2018; 9: 1275.
[Crossref] [Google Scholar] [Pubmed]
- Nantasenamat C, Prachayasittikul V. Maximizing computational tools for successful drug discovery. Expert Opin Drug Discov. 2015; 10(4): 321-329.
[Crossref] [Google Scholar] [Pubmed]
- Hajimahdi Z, Ranjbar A, Suratgar AA, Zarghi A. QSAR Study on anti-HIV-1 activity of 4-oxo-1, 4-dihydroquinoline and 4-oxo-4H-pyrido [1, 2-a] pyrimidine derivatives using SW-MLR, artificial neural network and filtering methods. Iran J Pharm Res. 2015; 14(Suppl): 69.
[Google Scholar] [Pubmed]
- Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science. 1983; 220(4599): 868-871.
[Crossref] [Google Scholar] [Pubmed]
- Palella Jr FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N Engl J Med. 1998; 338(13): 853-860.
[Crossref] [Google Scholar] [Pubmed]
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020; 583(7816): 459-468.
[Crossref] [Google Scholar] [Pubmed]
- Hajimahdi Z, Zabihollahi R, Aghasadeghi MR, Zarghi A. Design, synthesis and docking studies of new 4-hydroxyquinoline-3-carbohydrazide derivatives as anti-HIV-1 agents. Drug Res. 2013; 63(04): 192-197.
[Crossref] [Google Scholar] [Pubmed]
- Ivanov J, Polshakov D, Kato-Weinstein J, Zhou Q, Li Y, Granet R, et al. Quantitative structure-activity relationship machine learning models and their applications for identifying viral 3CLpro-and RdRp-targeting compounds as potential therapeutics for COVID-19 and related viral infections. ACS Omega. 2020; 5(42): 27344-27358.
[Crossref] [Google Scholar] [Pubmed]
- Te Velthuis AJ. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci. 2014; 71: 4403-4420.
[Crossref] [Google Scholar] [Pubmed]
- Comprehensive ensemble in QSAR prediction for drug discovery
- Nguyen KT, Blum LC, van Deursen R, Reymond JL. Classification of organic molecules by molecular quantum numbers. ChemMedChem. 2009; 4(11): 1803-1805.
[Crossref] [Google Scholar] [Pubmed]
- Ogura K, Sato T, Yuki H, Honma T. Support vector machine model for hERG inhibitory activities based on the integrated hERG database using descriptor selection by NSGA-II. Sci Rep. 2019; 9(1): 12220.
- Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006; 27(8): 861-874.
Author Info
Helit Jain1*, Dhananjay Meshram1 and Sharav Desai22Department of Pharmaceutics, Sanjivani College of Pharmaceutical Education and Research, Maharashtra, India
Citation: Jain H: Introduction and Application of Quantitative Structure Activity Relationship: A Review
Received: 12-Jun-2023 Accepted: 07-Jul-2023 Published: 14-Jul-2023, DOI: 10.31858/0975-8453.14.7.465-473
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3