Research Article - (2022) Volume 13, Issue 12
Abstract
The AS3MTM may be the most significant protein for the methylation of chemical elements species. The transfer of methyl radical from S-adenosyl-L-methionine (AdoMet) to powerfulness arsenical (AsIII) is catalyzed by the AS3MTM that is critical for arsenic metabolism in humans. Since the AS3MTM genetic polymorphism is linked to arsenic resistance, the association between the Single Ester Polymorphism (SNP) and AS3MTM inorganic arsenic (iAs) metabolism is being studied. Additionally, we tend to compared chemical action properties of recombinant human AS3MTM and AS3MTM/M287T. In reaction S-adenosyl methionine, arsenite, or methyl arsonous acid (MAsIII) as substrates and endogenous reductants, together with Glutathione (GSH), a Thioredoxin enzyme (TR) system and Tris (2-carboxyethyl) Pesticide complex (TCEP). By victimization of either TR or Trx or NADPH (Nicotinamide Adenine Dinucleotide Phosphate) or TCEP, AS3MTM catalyzes the conversion of iAsIII to MAsIII then to methyl radical sonic acid (MAsV), dimethyl arsinous acid (DMAsIII), and diethyl arsinic acid (DMAsV). The Cys156 and CyS206 gift in similarity model forms the binding website for AsIII. Cys32 and Cys61 are linked by disulphide bond. The most important product in the initiative of methylation is MAsIII which remains sure to protein until it gets methylated. The product is a lot of hepatotoxic and more malignant neoplastic disease powerfulness methyl arsenicals; however, arsenic undergoes oxidation and reduction as enzyme-bound intermediates.
Keywords
Genetic mutation, Arsenic metabolism, AS3MTM, MAsIII, DMAsIII
Introduction
As humans get evaluated they get custom-made to the surrounding environment. The adaption happens i.e. mutation (Melnick JG and Parkin G, 2007; Hanikenne M, et al., 2008). Mutation allows organisms to metabolize toxic things (Najarro MA, et al., 2015). The peoples can still metabolize low level. Scientists found that due to more consumption of arsenic water, the body has developed a genetic ability to metabolize arsenic. Scientists found that peoples of Andes can do metabolism arsenic (Grey R, 2015).
Arsenic can come in body from drinking water sources such as inorganic arsenic (iAs) in AsIII or AsV i.e. arsenite or arsenate (Caceres DD, et al., 2005; Caceres DD, et al., 2010). The AsIII (Arsenite) species are AsO3 -, HAsO3 2−, H2AsO3 − and H3AsO3, while AsV (Arsenate) species are: AsO4III−, HAsO4 2−, H2AsO4 − and H3AsO4. Group I type compounds i.e. inorganic arsenic (IARC, 2004). The safe level of arsenic in drinking water is 10 μg/L as per WHO, 2003. High arsenic exposure can show effects like skin pigmentation, hyperkeratosis, and cancer of bladder, liver, and kidney which may cause deaths (Wu MM, et al., 1989; Tondel M, et al., 1999).
The arsenic metabolises by dual pathways, which are oxidative methylation and reductive methylation. During metabolism arsenic transformed to Methylarsenite (MAsIII), Dimenthyl arsenite (DMAsIII), and sometimes may be to Trimethyl arsine (TMAsIII) by the enzyme AsIII S-adenosylmethionine methyltransferase (SAM) (Thomas DJ and Rosen BP, 2013; Zhu YG, et al., 2014). In the gastrointestinal tract the Methylarsenite (MAsIII), Dimenthyl arsenite (DMAsIII) are get methylated to form Monomethylarsonic acid (MMA) and Dimethyl arsinic acid (DMA). MMA and DMA are less toxic than both MAsIII and DMAsIII so they are readily excreted through urine, where they get oxidized abiotically to MAsV and DMAsV (Tseng CH, 2007). When the MAsV and DMAsV levels increase in the urine is an indication of Arsenic related diseases (Antonelli R, et al., 2014). The distribution of arsenic metabolites in urine is 10%-30% iAs, 0%-11% MAs and 26%-30% DMAs, but this distribution can vary from individual to individual (Vahter M, 2002). Single Nucleotide Polymorphisms in the hAS3MT gene are linked. Most SNPs have little effect on health, however the M287T SNP in hAS3MT can cause cancer and skin issues (Valenzuela OL, et al., 2009; de Chaudhuri S, et al., 2008). When methylation of inorganic arsenic occurs it produces toxic compound than previous (Naranmandura H, et al., 2012). For example, one protective AS3MT haplotype is prevalent in indigenous tribes in Argentina they consumed arsenic toxic water for long period. The concentration of arsenic in water is 0.8 mg/L and little urine excretion of MAs (7.5%) and a greater percentage of DMAs (78%).
In this study, we reviewed the human genetic related to AS3MTM to find out ability of enzymes to metabolise arsenic and the effect of amino acid substitution on it. Methylation of arsenic helps to prevent death and sever conditions. For this study, we synthesized the hAS3MT gene by bacterial synthesis, which helps us to get pure AS3MTM for further study. Then we compare the properties of enzymes in between hAS3MT and AS3MTM by using AsIII ionized molecule, which allows us to relate the structural and enzymatic property of both. In this, we show that AS3MTM is essential for arsenic methylation capacity and present evidence that HGTs (Horizontal Gene Transformation) from prokaryotes to eukaryotes underlie adaptations to arsenic which is also known as mutation. In this study we discussed about how the animals get evaluated from last centuries (Dheeman DS, et al., 2014).
Materials and Methods
We declare that, all methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by a Pravara Rural College of Pharmacy, Pravara nagar. All experiments were performed in accordance with relevant guidelines and regulations.
Reagents
Tris-(2-carboxyethyl)-phosphine (0.5 M, pH 7) was prepared. MAsV was reduced to trivalent MAsIII usingNa2S2O3, Na2S2O5, and H2SO4 and adjusted to pH 6.5 with NaOH (Chen J, et al., 2014). High Performance Liquid Chromatography (HPLC) coupled to inductively coupled mass spectroscopy validated the identities of the reduction products (ICP-MS). The methylation substrates were the Glutathione (GSH) conjugates As(GS) and MAs(GS), which were made by incubating 1 mM AsIII or MAsV with a four-fold molar excess of GSH in degassed buffers under argon for 5 hours at 23°C (Marapakala K, et al., 2012).
Strains and growth conditions
For plasmid E. coli was used by Dheeman DS, et al., 2014. Bacterial growth was monitored by measuring the optical density at 600 nm (A600nm).
Human AS3MTM gene cloning of hAS3MT cDNA
Chemically synthesized hAS3MT matching to the sequence of the cDNA clone, which lacks the final nine residues of the hAS3MT sequence, with codon optimization for expression in E. coli and sub-cloned into the EcoRV site of pUC57-Kan-hAS3MT. The synthetic hAS3MT gene was cloned into expression vector pMAL-c2x as an EcoRI/SalI digest from pUC57-KanhAS3MT, resulting in a fusion with the maltose-binding protein gene at the 5′ end and eight histidine residues at the 3′ end of the genomic sequence. The forward primer 5′-CCAGCCATGGCTGCACTTCGTGACGCTGAGA-3′ (NcoI site highlighted) and reverse primer 5′-CCTAGTCGACTCCAGCAGCATCAGGGACACATC-3′ were used to amplify the 1.1 kb fragment using PCR (SalI site underlined) (Dheeman DS, et al., 2014; Wood TC, et al., 2006).
Constructing mutation
Site-directed mutagenesis used to create mutations in the AS3MT gene. The conserved Cys32, Cys61, Cys156, and CyS206 residues were altered to serine codons, resulting in seven single-cysteine mutants of the synthetic hAS3MT. Commercial DNA sequencing verified each hAS3MT mutation (Sambrook J, et al., 1989).
Expression and purification of protein
By using the Ni-NTA chromatography Wild-type AS3MTM (87 837 Da) and variant enzymes are purified by using E. coli (Dheeman DS, et al., 2014). Cells carrying the plasmid pET41a-hAS3MT were grown at 37°C in 1 L of Luria Broth medium with 10 gm of tryptone, 5 gm of yeast extract, and 10 gm of NaCl per litre containing 50 g/mL Kanamycin for 3 hours before induction with 0.3 mM Isopropyl-D-1-thiogalactopyranoside (IPTG). After centrifuging the induced culture at 5000 rpm for 15 minutes at 4°C, it was suspended in 20 mL of buffer A containing 50 mM NaH2PO4 (pH 8.0), 1 mM TCEP, and 0.3 M NaCl, to which 10 mM imidazole was added. The cells were lysed in the presence of Di-isopropyl fluorophosphate using a press before being centrifuged at 35000 rpm for 1 hour. Then, apply the aforementioned solution (0.7 mL/min) to a Ni-NTA agarose column that has already been loaded with 5 column volumes of buffer. hAS3MT was then eluted (0.7 mL/min) with 8 column buffer containing 0.25 M imidazole after being column washed (1 mL/min) with 10 of buffer containing 20 mM imidazole. The Imidazole is then removed. As previously disclosed, natural hAS3MT was purified. Purify Thioredoxin (Trx) and Thioredoxin Reductase (TR) from E. coli BL21(DE3) bearing either pET14b-trxA or pET14b-taxi using Ni-NTA chromatography as stated above. Before use, all buffers were degassed (aliquoted) by bubbling with argon for 30 minutes (Dheeman DS, et al., 2014).
Metabolism of arsenic (AS3MTM)
There are two pathways by which the human body metabolize the arsenic compound i.e. Methylation by Oxidation and Reduction type of reaction.
The activity of AS3MTMs was checked at 37°C in a buffer of 50 mM NaH2PO4 pH 8 and 0.3 M NaCl. The chemicals in the assay are 5 mM GSH, 1 mM SAM, 10 μM Trx, 3 μM TR, and 0.3 mM NADPH, and the reactions were terminated by adding 10% (v/v) H2O2 to oxidize all arsenic species. Centrifugation using a 3 kDa cut-off Amicon ultra-filter was used to remove denatured protein and then analysed by using HPLC.
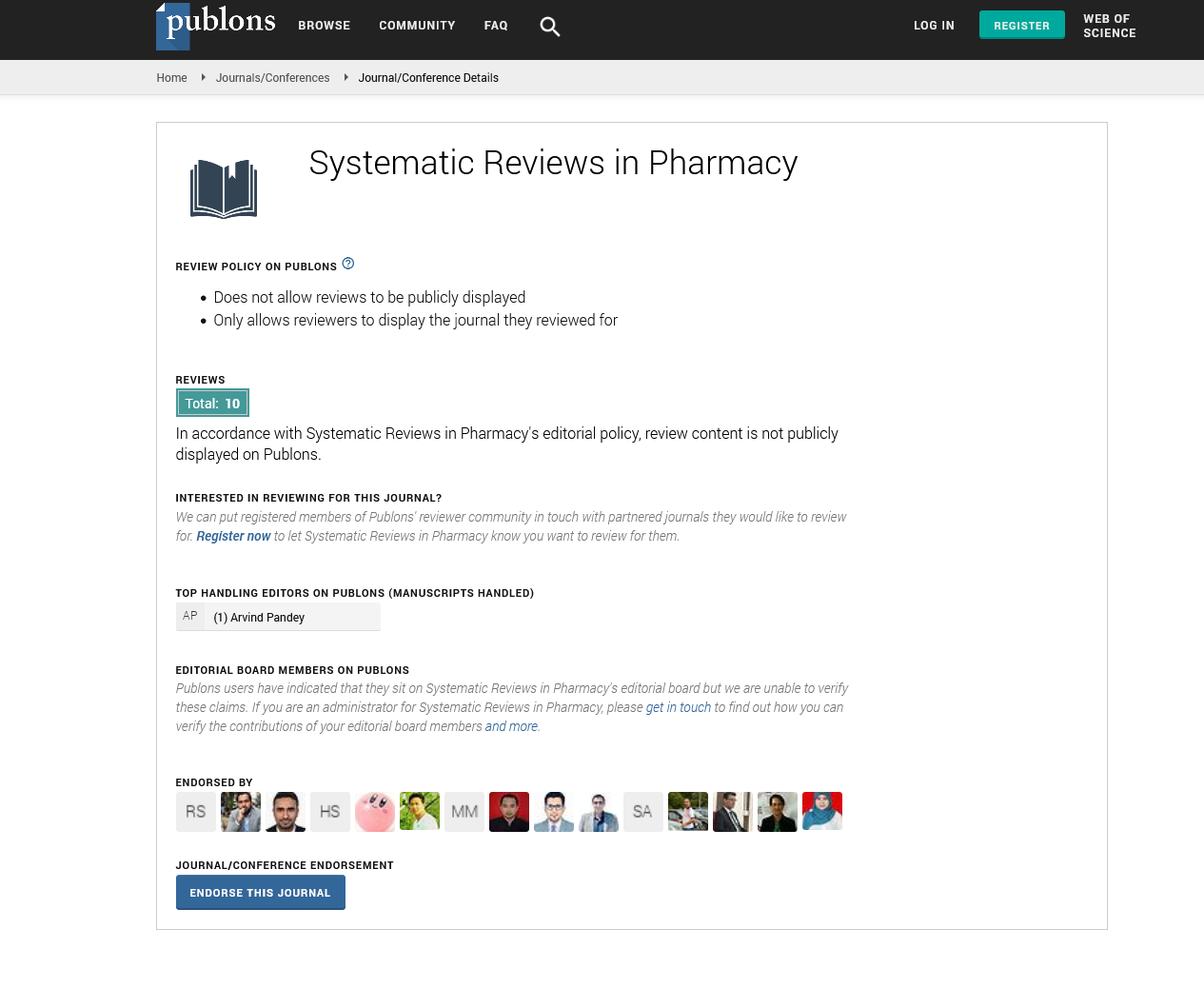
Oxidative methylation: This process is also called as bioactive process. This pathway is given by Cullen and Reimer. By the combination of Oxidative methylation, arsenate (AsV) is changed to Dimethylarsinous acid (DMAIII) (Figure 1a). Then Arsenate (AsV) gets converted into Arsenite (AsIII) following to Monomethylarsonic acid (MMAV) then it gets converted into Mono-methylarsonous acid (MMAIII). Then MMAIII converts into Dimethylarsinic acid (DMAV) and finally it forms Dimethylarsinous acid (DMAIII). We can’t explain the complete metabolism process because of the detection of DMAV arsenic which occurs in a major amount in human urine. Because the toxicity of MMAV and DMAV is substantially lower than that of iAs, methylation is thought to be a detoxification step for iAs. According to several recent investigations, MMAIII or DMAIII are more cytotoxic and genotoxic than iAs (Cullen WR, Reimer KJ, 1989; Petrick JS, et al., 2000). We noticed that if we cannot add H2O2 it allow us to determine trivalent arsenicals.
Figure 1a: Oxidative methylation of arsenic
Reductive methylation: This pathway of arsenic was proposed by Hayakawa T, et al., 2005. In this pathway, trivalent arsenicals are conjugated with glutathione (GSH) and then they get methylated. In the first step, AsIII changes to AsIIIGS3 then MMA+3GS2 is formed then later it gets reduced to Dimethylarsenoglutathione (DMAIIIGS) (Figure 1b). Then correspondingly the MMA+3GS2 and DMAIIIGS are get oxidized to MMAV and DMAV. We investigated the renal metabolites and hepatic metabolites after giving the arsenic intrAVenously to the mice (0.5 mg/kg body weight), then we observed that when a trivalent species (AsIII) of arsenic binds to a thiol group (R-SH) present in proteins. Then the protein-arsenical complex detaches from a parent protein and forms conjugation with Glutathione (GSH) to form AsIII(GS)3 or MMAIII(GS)2 or DMAIII(GS). Hence it is found that during reductive methylation MMAV and DMAV are the end products. But in this pathway, DMAV is in the major amount present in urine called as detoxification (Naranmandura H, et al., 2006).
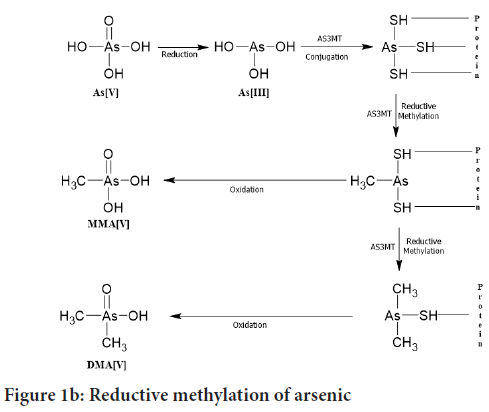
Figure 1b: Reductive methylation of arsenic
At the point when iAs is methylated through Oxidative and Reductive Methylation, the AS3MTM quality assumes a basic part. AS3MTM is an S-adenosyl-L-methionine-subordinate compound that can methylate trivalent arsenicals (Wood TC, et al., 2006). The human AS3MTM gene is 32 kb long and has 11 exons. A variety of genetic variations SNP. A VNTR (Variable Number of Tandem Repeats) is a spot in DNA where a short nucleotide sequence is organized (Wood TC, et al., 2006). When AS3MTM methylates inorganic arsenic, it can cause oxidative DNA damage and enhance their carcinogenicity.
Assays of arsenic methylation
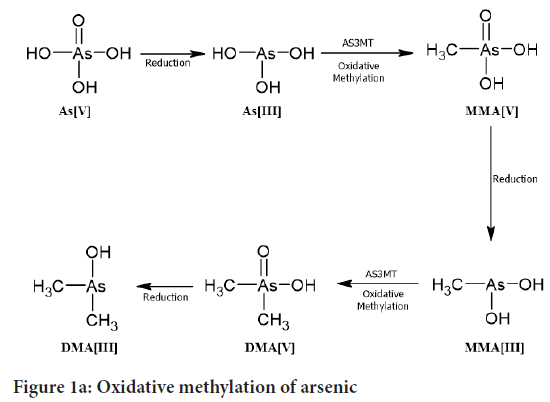
To assay measurement of conversion of SAM to S-adenosyl homocysteine (SAH) EPI generous Methyltransferase Assay kit is used where the Time-Resolved Förster Resonance Energy Transfer (TR-FRET) is used (Dong H, et al., 2015). The test was completed utilizing a 384-well microtiter plate in a cushion comprising of 50 mM NaH2PO4 (pH 8.0), containing 0.3 M NaCl, 1 μM cleaned hAS3MT, 0.5 mM GSH, 1 μM Trx, 0.3 μM TR and 0.03 mM NADPH and 10 μM of As(GS) or MAs(GS). Then we added the SAM at 10 μM. The emission was 665:620 nm for determine Homogeneous Time-Resolved Fluorescence (HTRF). The concentration calculated as given in Figure 2 (Dong H, et al., 2015).
Figure 2: Assays of arsenic methylation
For measurement, (HPLC) and for arsenic concentration Inductively Coupled Plasma Mass Spectrometry (ICP-MS) was used (Dheeman DS, et al., 2014). Then added the SAM to at 37°C. To recuperate the entirety of the arsenic, the responses were ended by the expansion of H2O2 at 10% (v/v) last fixation, which additionally oxidizes all arsenicals, so the items will be named MAs and DMAs. Speciation of arsenic in the still up in the air by HPLC with a C18 300A opposite stage section with the arsenic focus estimated by ICP-MS utilizing an ELAN 9000 ICP-MS. AsIII, MMAIII, DMAV, MAV, and AsV were utilized at 1 μM as principles.
E. coli cells expressing the genotype and mutants of the hAS3MT and hence, we carried both methylation processes on them. The cells were grown for 12 hours at 37°C in a 2 mL liquid broth medium of 0.3 mM Isopropyl-D-1-thiogalactopyranoside (IPTG), 100 g/mL kanamycin, and 10 M of AsIII or 2 M MMAIII or both was used. The cells were extracted, washed, and suspended in ST-1 media with 2 M MMAIII before being cultured at 37°C for 3 hours (Gill SC and von Hippel PH, 1989). Arsenicals were spectated by HPLC using a C18 reverse phase column, and the quantity of arsenic was calculated by Inductively Coupled Plasma Mass Spectroscopy (ICP-MS).
hAS3MT structure homology model with polymorphic residues
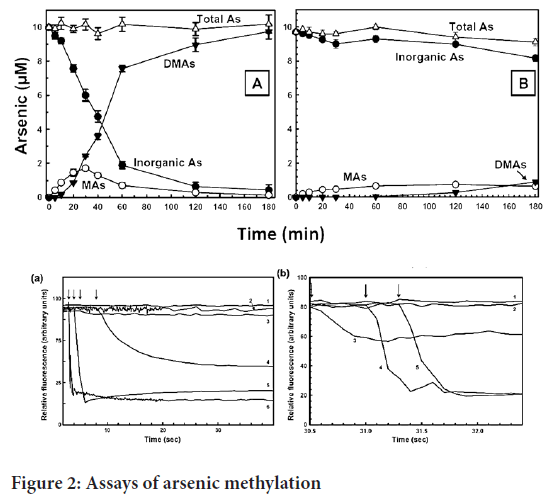
Using a fully automated protein structure homology modelling system, a homology model of hAS3MT was generated from residues on the structure of PhAsIII, which is confined to CmArsM. We utilize the PATCHDOCK server to find SAM’s position in the model. The AsIII bound structure of CmArsM (Ajees AA, et al., 2012) was overlaid on the found hAS3MT model using SAM. We used a visual technique to get the arsenic atom in the AsIII binding site of hAS3MT (Figure 3). The human AS3MT model structure is depicted in a cartoon diagram with a tan colour scheme.
Figure 3: Homology structural model of human AS3MTM
The relationship between arsenic methylation and genotypes in human AS3MTM
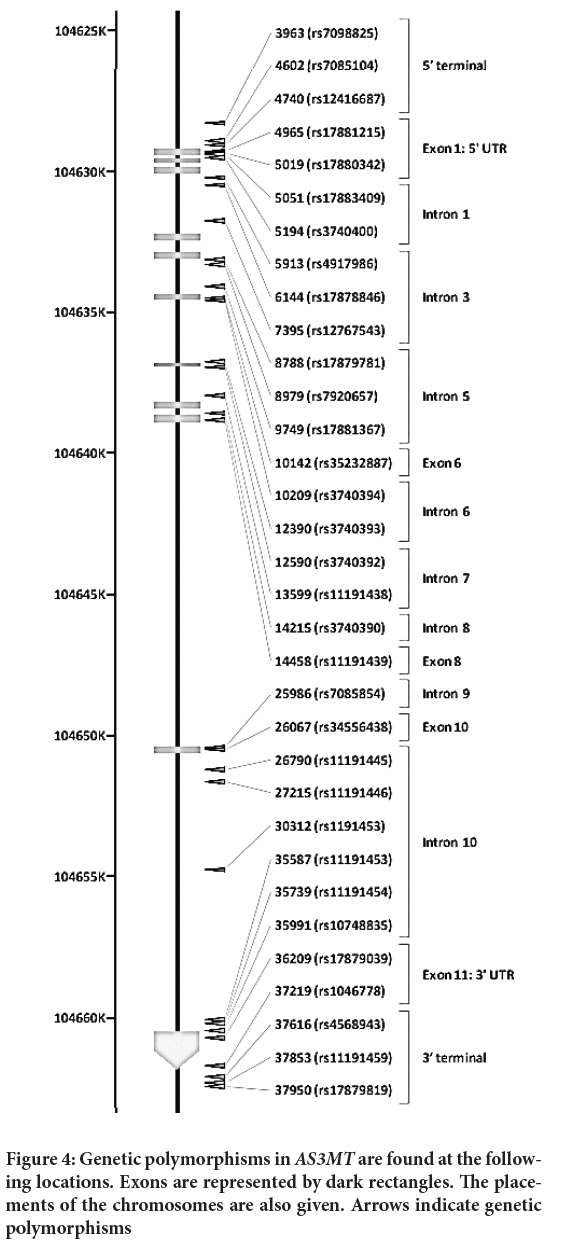
In this study, we employ SNP, which stands for polymorphism identification number related to the consensus sequence site (AY817668), with the first base of the consensus number 1 and dbSNP rs# cluster id (Tseng CH, 2009). Figure 4 depicts the chromosomal positions of genetic polymorphisms in AS3MTM.
Figure 4: Genetic polymorphisms in AS3MTM are found at the following locations. Exons are represented by dark rectangles. The placements of the chromosomes are also given. Arrows indicate genetic polymorphisms
Out of all SNPs, three of them has non-synonymous exon region, which is Arg173Trp, Met287Thr, and Thr306Ile. When these AS3MTM-expressing cells are treated with 12.5 nM AsIII, the Met287. During our study, we discovered that repeated sequences of 5’-UTR VNTR influences the transcriptional expression of gene. According to the findings of this study, polymorphisms in AS3MTM lead to individual variability in AS3MTM expression and function, as well as variance in the risk of arsenic-dependent carcinogenesis.
Animal case study
This study was conducted on the selected animals (mice’s) are treated with arsenic containing water in different concentrations (González-Martínez F, et al., 2018).
Collection of different samples
Groundwater sample collection: We collected the water from 15 random groundwater wells in the polythene bottle which is previously treated with 25% HNO3 for 3 hours. Then the bottles are washed with water. After sample were kept under refrigerated at the laboratory.
Blood sample collection: Blood samples from subjects were collected by venous puncture using lead-free vacutainer tubes containing EDTA (Ethylenediamine tetraacetic acid) as an anticoagulant. The blood was collected from all subjects and transferred to laboratory for study. DNA was extracted through the HP-PCR (High Pure-Polymerase Chain Reaction).
Collection of a urine sample: Approximately 15 ml of urine sample is collected bottle and the bottle is cleaned as mentioned in Groundwater is also collected. The sample was freeze to prevent oxidation. Then sample was filtered to remove unwanted waste.
Risk assessment
The following calculation was used to assess human exposure to arsenic in groundwater using the Lifetime Average Daily Dose (LADD), which is the amount of daily arsenic exposed from one or more sources and is given in g of arsenic per kilogramme body weight per day (g/kg/day):
LADD=C × IR × ED × EF/BW × AT
Where,
C-Arsenic concentration in water (μg/L),
IR-The water intake rate per day (L/day),
ED-Exposure duration (years),
EF-The exposure frequency (days/year),
Kg-The body weight/Kg,
AT-The average time (day)
By the LADD value we determined the Hazard Quotient (HQ) by the following equation;
HQ=LADD/RFD
Where,
RfD is reference dose for arsenic is 10 μg/kg/day as given by WHO for India to avoid non-cancerous outcomes such as hyperpigmentation, keratosis, and possible vascular complications.
Determination of groundwater quality and arsenic
It was necessary to check turbidity, pH, conductivity, temperature, dissolved oxygen, chloride, fluoride, nitrite, nitrate, magnesium, and other physiochemical parameters. In the laboratory, chloride (mg/L) and fluoride (mg/L) were measured using a Benchtop Multiparameter pH/ISE with the appropriate ion-selective electrodes (Marchiset-Ferlay N, et al., 2012).
Urinary Arsenic Species (UAs) determination by instrumental analysis: Using HPLC-HG-AFS, the urinary arsenic species (UAS) (AsIII, AsV, MMA, and DMA) were determined (High-Performance Liquid Chromatography-Hydride Generation-Atomic Fluorescence Spectrometry). A Hamilton PRP-X100 anion-exchange column with a diameter of 250 mm aqueous buffer KH2PO4 or K2HPO4 with a pH of 5.8 is used as the mobile phase. The flow rate was 1.0 mL/min. To find out total urinary arsenic, the urine sample is subjected within HNO3 and HClO4 to convert arsenic to inorganic arsenic (iAs). Finally, the HG-AFS method is employed to calculate UAs (Hydride Generation-Atomic Fluorescence Spectrometry) (Meza MM, et al., 2004).
Quality assurance for arsenical: Limit of Detection (LOD) and Limit of Quantification (LOQ) were employed to detect arsenic in water, yielding values of 0.7 g/L and 1.2 g/L, respectively. The concentration of arsenic in urine is measured. We reported 106.22 g/L of total iAs, which is the sum of the AsIII and AsV. We also used HPLC-HG-AFS to verify the recovery of arsenic species, yielding a total of 107.8 2.4 g/L, which matched to 99.5 2.1 g/L of AsV and 8.3 0.3 g/L of DMA. The following were the Urinary Arsenic Species (UAS) Limits of Detection (LOD): AsIII is 0.17 g/L, AsV is 0.38 g/L, MMA is 0.30 g/L, and DMA is 0.45 g/L.
Statistical analysis
Because arsenic concentrations in groundwater and urinary arsenic species do not have a normal distribution. The genotype distributions of GSTP1-rs1695, GSTO2-rs156697, and AS3MTM-rs3740400 were measured using the Hardy-Weinberg Equilibrium (HWE). Allelic frequencies were obtained by dividing the frequencies of heterozygous and homozygous alleles by the total number of allelic variants. We split the total number of individuals into two groups: Those with a low daily dosage (LADD 0.3 g/ kg/day) and those with a high daily dose (LADD>0.3 g/kg/day). We do the comparison between low and high exposure doses. To understand the differences between low and high intake of toxics, an effect size test was performed (Hernández A, et al., 2008). The polymorphisms and LADD were used as independent factors in the study, while urine arsenic species were used as dependent variables. The study included genetic dominant models (heterozygous+homozygous genotype) as well as possible confounders (age, BMI, smoking history, and lifestyle). With a stronger biological sense, dominant models over Potential Confounders model are explored in the research population. We used various factors to analyse the multi-collinearity of independent variables (VIF) (Lesseur C, et al., 2012).
Results
Methylation of arsenic is conversion from product to substrate (Huang JH, et al., 2007; Huang YL, et al., 2009). The ratio of MAs/iAs is the primary methylation index, While the ratio of DMAs/MAs is the Secondary Methylation Index (SMI). The SMI is primarily used to assess methylation capability in persons exposed to inorganic arsenic (Chen GQ, et al., 2003). We evaluated the methylation index of wild-type hAS3MT to that of eight polymorphic enzymes in this work. SMI and PMI (Primary Methylation Index) were found to be lower in eight polymorphic enzymes. The greatest value of the DMAs/MAs ratio in wild-type enzymes is 2.3 0.3. M287T, R251H, and T306I SMI values (about 1.2) were lower than the wild-type enzyme but higher than the other SNPs, while the SMI values of H51R, I136T, and R173W enzymes were roughly 0.45. This research demonstrates that the eight non-synonymous missense variations of hAS3MT had a decreased arsenic methylation capacity when compared, implying that there is variance in arsenic methylation from individual to individual, which may raise the risk of arsenic-related disorders.
Discussion
During the transformation of arsenic, the conversion of inorganic arsenic to Methylarsonic acid and dimethylarsinic acid is the most important step (Abernathy CO, et al., 2012). On basis of metabolism processes of arsenic we were curious to find that is it is suitable to inhibit cancer or not (Sanz MA, et al., 2005).
The main step is to give annotation of the AS3MTM gene, resulting in several differences from the current “provisional” NCBI annotation. During the re-sequencing, we identified 27 polymorphisms, including three non-synonymous cSNPs and a VNTR. For allozymes, Trp173 and Ile306, levels of enzyme activity and immune-reactive protein were strikingly decreased when compared with the WT allozyme (Wang L, et al., 2003; Thomae BA, et al., 2003; Shield AJ, et al., 2004).
All of the substituted residues are on the surface of the protein except for Thr306, which is buried inside the enzyme, so a T306I substitution is disrupt the structure (Figure 3). The second methylation step is reduced when M287T SNP is occur (Agusa T, et al., 2011). Met287 is located on the surface of AS3MTM where the molecular inhibitors bind (Dong H, et al., 2015). We also see that when M287T substitutes the binding site it retards the allosteric conformational change and reduces the rate of methylation of this variant. This property is observed in individuals with the M287T polymorphism epidemiological studies.
During the stability study in temperature, we found that the variants denature between 4 to 20 fold faster than wild-type h AS3MTM. The protective AS3MTM polymorphisms are located outside of the coding sequence regulatory elements.
Conclusion
The gene-gene interactions AS3MTM*GSTM1 and GSTO2*GSTP1 were discovered to be possible regulators of urinary arsenic metabolites, increasing MMA and decreasing DMA, in this work. A synergistic effect of these polymorphisms and age, LADD of arsenic, and alcohol use may also alter a significant fraction of the population’s arsenic individual metabolic capacity. Despite some discrepancies between genotypes and metabolism in human case studies, we discovered that two SNPs, AS3MTM 12390 (rs3740393) in intron and 14458 (rs11191439, Met287Thr) in exon, vary across all nations, indicating that SNPs may be ethnically independent polymorphisms, but they can affect arsenic methylation. Argentina’s population has a more proportion of DMA and a lower DM. This different distribution may have led to the findings that Argentina’s population had a higher percentage of DMA and a lower percentage of MMA in the urine when compared to other research. It’s worth looking into if the genotype distribution of AS3MTM 12390 (rs3740393) is unique to this group (Argentinean Andes) and how this unique SNP selection happened.
Declarations
Acknowledgements
We thank Prof. Mayur Bhosale for assistance with genetics, and Dr. Sanjay Bhawar, Principal, Pravara Rural College of Pharmacy, Pravaranagar for comments that greatly improved the manuscript.
Ethics statement
This is an observational study. The Pravara Research Ethics Committee has confirmed that no ethical approval is required.
Consent to participate (Ethics)
Informed consent was obtained from all individual participants included in the study.
Consent to publish (Ethics)
The participant has consented to the submission of the article to the journal.
Author contributions
“All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Pratik V. Malvade, Mayur S. Bhosale, and Sayli R. Chavan. The first draft of the manuscript was written by Pratik V. Malvade and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.”
Availability of data and material
We do not wish to share our data before we have thoroughly analyzed it. All data sources described in the study are directed at the corresponding author.
Data availability statement
In this article, data sharing not applicable as no datasets were generated or analyzed during the current study.
References
- Melnick JG, Parkin G. Cleaving mercury-alkyl bonds: A functional model for mercury detoxification by MerB. Science. 2007; 317(5835): 225-227.
[Crossref] [Google Scholar] [Pubmed]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, et al. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008; 453(7193): 391-395.
[Crossref] [Google Scholar] [Pubmed]
- Najarro MA, Hackett JL, Smith BR, Highfill CA, King EG, Long AD, et al. Identifying loci contributing to natural variation in xenobiotic resistance in Drosophila. PLoS Genet. 2015; 11(11): e1005663.
[Crossref] [Google Scholar] [Pubmed]
- Grey R. The people who can eat arsenic: Remote village in Andes found to have developed tolerance to toxic chemical. Mail Online. 2015.
- Caceres DD, Pino P, Montesinos N, Atalah E, Amigo H, Loomis D. Exposure to inorganic arsenic in drinking water and total urinary arsenic concentration in a Chilean population. Environ Res. 2005; 98(2): 151-159.
[Crossref] [Google Scholar] [Pubmed]
- Caceres DD, Werlinger F, Orellana M, Jara M, Rocha R, Alvarado SA, et al. Polymorphism of Glutathione S-transferase (GST) variants and its effect on distribution of urinary arsenic species in people exposed to low inorganic arsenic in tap water: An exploratory study. Arch Environ Occup Health. 2010; 65(3): 140-147.
[Crossref] [Google Scholar] [Pubmed]
- IARC (Working Group on the Evaluation of Carcinogenic Risks to Humans). Some drinking-water disinfectants and contaminants, including arsenic. IARC Monogr Eval Carcinog Risks Hum. 2004; 84: 1-477.
[Google Scholar] [Pubmed]
- WHO. Arsenic in drinking-water: Background document for development of WHO guidelines for drinking-water quality. World Health Organization; 2003.
- Wu MM, Kuo TL, Hwang YH, Chen CJ. Dose-response relation between arsenic concentration in well water and mortality from cancers and vascular diseases. Am J Epidemiol. 1989; 130(6): 1123-1132.
[Crossref] [Google Scholar] [Pubmed]
- Tondel M, Rahman M, Magnuson A, Chowdhury IA, Faruquee MH, Ahmad SA. The relationship of arsenic levels in drinking water and the prevalence rate of skin lesions in Bangladesh. Environ Health Perspect. 1999; 107(9): 727-729.
[Crossref] [Google Scholar] [Pubmed]
- Thomas DJ, Rosen BP. Arsenic methyltransferases. Encyclopedia of Metalloproteins. 2013; 138-143.
- Zhu YG, Yoshinaga M, Zhao FJ, Rosen BP. Earth abides arsenic biotransformations. Annu Rev Earth Planet Sci. 2014; 42: 443.
[Crossref] [Google Scholar] [Pubmed]
- Tseng CH. Arsenic methylation, urinary arsenic metabolites and human diseases: Current perspective. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2007; 25(1): 1-22.
[Crossref] [Google Scholar] [Pubmed]
- Antonelli R, Shao K, Thomas DJ, Sams II R, Cowden J. AS3MT, GSTO, and PNP polymorphisms: Impact on arsenic methylation and implications for disease susceptibility. Environ Res. 2014; 132: 156-167.
[Crossref] [Google Scholar] [Pubmed]
- Vahter M. Mechanisms of arsenic biotransformation. Toxicology. 2002; 181: 211-217.
[Crossref] [Google Scholar] [Pubmed]
- Valenzuela OL, Drobná Z, Hernández-Castellanos E, Sánchez-Peña LC, García-Vargas GG, Borja-Aburto VH, et al. Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol Appl Pharmacol. 2009; 239(2): 200-207.
[Crossref] [Google Scholar] [Pubmed]
- de Chaudhuri S, Ghosh P, Sarma N, Majumdar P, Sau TJ, Basu S, et al. Genetic variants associated with arsenic susceptibility: Study of purine nucleoside phosphorylase, arsenic (+ 3) methyltransferase, and glutathione S-transferase omega genes. Environ Health Perspect. 2008; 116(4): 501-505.
[Crossref] [Google Scholar] [Pubmed]
- Naranmandura H, Chen X, Tanaka M, Wang WW, Rehman K, Xu S, et al. Release of apoptotic cytochrome C from mitochondria by dimethylarsinous acid occurs through interaction with voltage-dependent anion channel in vitro. Toxicol Sci. 2012; 128(1): 137-146.
[Crossref] [Google Scholar] [Pubmed]
- Dheeman DS, Packianathan C, Pillai JK, Rosen BP. Pathway of human AS3MT arsenic methylation. Chem Res Toxicol. 2014; 27(11): 1979-1989.
[Crossref] [Google Scholar] [Pubmed]
- Chen J, Sun S, Li CZ, Zhu YG, Rosen BP. Biosensor for organoarsenical herbicides and growth promoters. Environ Sci Technol. 2014; 48(2): 1141-1147.
[Crossref] [Google Scholar] [Pubmed]
- Marapakala K, Qin J, Rosen BP. Identification of catalytic residues in the As (III) S-adenosylmethionine methyltransferase. Biochemistry. 2012; 51(5): 944-9451.
[Crossref] [Google Scholar] [Pubmed]
- Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, et al. Human arsenic methyltransferase (AS3MT) pharmacogenetics: Gene resequencing and functional genomics studies. J Biol Chem. 2006; 281(11): 7364-7373.
[Crossref] [Google Scholar] [Pubmed]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manule. Cold Spring Harbor Laboratory. 1989.
- Cullen WR, Reimer KJ. Arsenic speciation in the environment. Chem Rev. 1989; 89: 713-764.
- Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE, Aposhian HV. Monomethylarsonous acid (MMAIII) is more toxic than arsenite in Chang human hepatocytes. Toxicol Appl Pharmacol. 2000; 163(2): 203-207.
[Crossref] [Google Scholar] [Pubmed]
- Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: Arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005; 79(4): 183-191.
[Crossref] [Google Scholar] [Pubmed]
- Naranmandura H, Suzuki N, Suzuki KT. Trivalent arsenicals are bound to proteins during reductive methylation. Chem Res Toxicol. 2006; 19(8): 1010-1018.
[Crossref] [Google Scholar] [Pubmed]
- Dong H, Xu W, Pillai JK, Packianathan C, Rosen BP. High-throughput screening-compatible assays of As (III) S-adenosylmethionine methyltransferase activity. Anal Biochem. 2015; 480: 67-73.
[Crossref] [Google Scholar] [Pubmed]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989; 182(2): 319-326.
[Crossref] [Google Scholar] [Pubmed]
- Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP. Structure of an As (III) S-adenosylmethionine methyltransferase: Insights into the mechanism of arsenic biotransformation. Biochemistry. 2012; 51(27): 5476-5485.
[Crossref] [Google Scholar] [Pubmed]
- Tseng CH. A review on environmental factors regulating arsenic methylation in humans. Toxicol Appl Pharmacol. 2009; 235(3): 338-350.
[Crossref] [Google Scholar] [Pubmed]
- González-Martínez F, Sánchez-Rodas D, Cáceres DD, Martínez MF, Quiñones LA, Johnson-Restrepo B, et al. Arsenic exposure, profiles of urinary arsenic species, and polymorphism effects of glutathione-s-transferase and metallothioneins. Chemosphere. 2018; 212: 927-936.
[Crossref] [Google Scholar] [Pubmed]
- Marchiset-Ferlay N, Savanovitch C, Sauvant-Rochat MP. What is the best biomarker to assess arsenic exposure via drinking water?. Environ Int. 2012; 39(1): 150-171.
[Crossref] [Google Scholar] [Pubmed]
- Meza MM, Kopplin MJ, Burgess JL, Gandolfi AJ. Arsenic drinking water exposure and urinary excretion among adults in the Yaqui Valley, Sonora, Mexico. Environ Res. 2004; 96(2): 119-126.
[Crossref] [Google Scholar] [Pubmed]
- Hernández A, Xamena N, Sekaran C, Tokunaga H, Sampayo-Reyes A, Quinteros D, et al. High arsenic metabolic efficiency in AS3MT287Thr allele carriers. Pharmacogenet Genomics. 2008; 18(4): 349-355.
[Crossref] [Google Scholar] [Pubmed]
- Lesseur C, Gilbert-Diamond D, Andrew AS, Ekstrom RM, Li Z, Kelsey KT, et al. A case-control study of polymorphisms in xenobiotic and arsenic metabolism genes and arsenic-related bladder cancer in New Hampshire. Toxicol Lett. 2012; 210(1): 100-106.
[Crossref] [Google Scholar] [Pubmed]
- Huang JH, Scherr F, Matzner E. Demethylation of dimethylarsinic acid and arsenobetaine in different organic soils. Water Air Soil Pollut. 2007; 182(1): 31-41.
- Huang YL, Hsueh YM, Huang YK, Yip PK, Yang MH, Chen CJ. Urinary arsenic methylation capability and carotid atherosclerosis risk in subjects living in arsenicosis-hyperendemic areas in southwestern Taiwan. Sci Total Environ. 2009; 407(8): 2608-2614.
[Crossref] [Google Scholar] [Pubmed]
- Chen GQ, Zhou L, Styblo M, Walton F, Jing Y, Weinberg R, et al. Methylated metabolites of arsenic trioxide are more potent than arsenic trioxide as apoptotic but not differentiation inducers in leukemia and lymphoma cells. Cancer Res. 2003; 63(8): 1853-1859.
[Google Scholar] [Pubmed]
- Abernathy CO, Calderon RL, Chappell WR. Arsenic: Exposure and health effects. Springer Science and Business Media. 2012.
- Sanz MA, Fenaux P, Coco FL. Arsenic trioxide in the treatment of acute promyelocytic leukemia: A review of current evidence. Haematologica. 2005; 90(9): 1231-1235.
[Crossref] [Google Scholar] [Pubmed]
- Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: Chaperone protein association and allozyme degradation. Pharmacogenetics. 2003; 13(9): 555-564.
[Crossref] [Google Scholar] [Pubmed]
- Thomae BA, Rifki OF, Theobald MA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catecholamine sulfotransferase (SULT1A3) pharmacogenetics: Functional genetic polymorphism. J Neurochem. 2003; 87(4): 809-819.
[Crossref] [Google Scholar] [Pubmed]
- Shield AJ, Thomae BA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catechol O-methyltransferase genetic variation: Gene resequencing and functional characterization of variant allozymes. Mol psychiatry. 2004; 9(2): 151-160.
[Crossref] [Google Scholar] [Pubmed]
- Agusa T, Fujihara J, Takeshita H, Iwata H. Individual variations in inorganic arsenic metabolism associated with AS3MT genetic polymorphisms. Int J Mol Sci. 2011; 12(4): 2351-2382.
[Crossref] [Google Scholar] [Pubmed]
- Dong H, Madegowda M, Nefzi A, Houghten RA, Giulianotti MA, Rosen BP. Identification of small molecule inhibitors of human As (III) S-adenosylmethionine methyltransferase (AS3MT). Chem Res Toxicol. 2015; 28(12): 2419-2425.
[Crossref] [Google Scholar] [Pubmed]
Author Info
Pratik Vijay Malvade*, Mayur S Bhosale, Sayli R Chavan and Dhanashri B BhagatCitation: Malvade PV: Metabolism of Arsenic in Human by AS3MT Gene
Received: 01-Nov-2022 Accepted: 25-Nov-2022 Published: 02-Dec-2022, DOI: : 10.31858/0975-8453.13.12.819-825
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3