Research Article - (2022) Volume 13, Issue 11
Abstract
Background: Bacterial infections are the commonest in both community and healthcare settings. Emergency of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae has contributed to poor clinical outcomes. More efforts regarding antibiotic resistance have been dedicated to clinical settings and we do not know the extent of the catastrophe in community settings. We aimed at determining the burden, antimicrobial susceptibility patterns and molecular characteristics of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae in agro-pastoral communities of Mbarara district, South Western Uganda.
Methods: A laboratory based descriptive cross-sectional study was carried out among Enterobacteriaceae isolated from outpatients presenting with signs and symptoms of Urinary Tract Infections. Urine samples were delivered to Microbiology Laboratory of Mbarara University of Science and Technology for culture, identification, testing for ESBL production and Antibiotic Susceptibility Testing. Molecular characterization of ESBL producing Enterobacteriaceae was carried out at Medical and Molecular Laboratories Limited of Makerere University.
Results: A total of 88 Enterobacteriaceae fulfilling the inclusion criteria were considered into the study. Escherichia coli 70.45% and Klebsiella pneumoniae 13.64% were the most isolated followed by Klebsiella oxytoca, Proteus mirabilis and Enterobacter aerogenes at 10.23%, 3.41% and 2.27% respectively. The production of ESBL was observed at 23.86%. Generally, high resistance rates were observed against Ampicillin 100%, Cefepime 100%, Aztreonam 95.24%, Nalidixic acid 90.48%, Ciprofloxacin 85.71% and Amoxicillin/clavulanate 80.95%. High rates of sensitivity were observed to Meropenem 95.24%, Imipenem 95.24%, Amikacin 95.24%, Gentamycin 90.48%, Cefoxitin 76.19% Piperacillin/tazobactam 80.95% and Nitrofurantoin 66. 67%. Multi-Drug Resistance (MDR) was observed at 85.71%. The most prevalent genes in ESBL producing Enterobacteriaceae were CTX-MU (Cefotaxime-Munich) 46.7%, TEM 30.00% and SHV (Sulfhydryl Reagent Variable) 23.3%.
Conclusion: We demonstrated high prevalence, antibiotic resistance rates among Extended Spectrum Beta-Lactamase producing Enterobacteriaceae in the community. We recommend more community ESBL related studies and a One Health Approach to guide public health interventions.
Keywords
Enterobacteriaceae, Extended Spectrum Beta-Lactamase (ESBL), Bacterial infections
Introduction
Resistance of pathogenic bacteria to available antimicrobial agents has become a worldwide public health concern requiring an appropriate intervention (Laxminarayan R, et al., 2013). This has made the treatment of bacterial infections more difficult. The burden of Antimicrobial Resistance varies greatly across continents worldwide. Resistance to antimicrobial agents in middle income countries is generally higher than that in high income countries, with general prevalence varying between 20%-70% (Klein EY, et al., 2019). To date, 52 countries are included in World Health Organization’s Global Antimicrobial Surveillance System with resistance reported between 8%-65% (WHO, 2018). In East Africa where Uganda is found, Antimicrobial Resistance (AMR) of Enterobacteriaceae is estimated between 46%-69% according to a situational analysis in the review of AMR in East Africa (Ampaire L, et al., 2016).
The global burden of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae is estimated to be between 25%-50% in infections and 20%-40% in healthy population (Frost I, et al., 2019) while in Uganda, the burden varies greatly according to the region and whether the study included inpatients only, outpatient only or both and is generally between 5%-62% (Kateregga JN, et al., 2015; Najjuka CF, et al., 2016).
Extended Spectrum Beta-Lactamase (ESBL) producing Enterobacteriaceae have become a challenge to patients, clinicians, public health professionals in healthcare facilities (Ben-Ami R, et al., 2006). Community acquired infections due to ESBL producing organisms have been described in some countries including Spain, Israel, the United Kingdom and Canada and these are usually Urinary Tract Infections (UTIs).
Most bacterial infections are treated with BetaLactam antibiotics especially in rural areas of Uganda which include urinary tract infections, bacteremia, respiratory infections and sepsis (Andrew B, et al., 2017). However, most Enterobacteriaceae produce β-lactamase enzymes so as to overcome the action of β-lactam antibiotics (Shaikh S, et al., 2015). Extended-Spectrum Beta-Lactamases (ESBLs) are the type of β-lactamase enzymes produced by most members in the family of Enterobacteriaceae (Brolund A, 2014) enabling the bacteria to resist penicillins, cephalosporins up to third generation and aztreonam by hydrolysis (Pagani L, et al., 2002) and the genes responsible for production of these enzymes are encoded on mobile genetic elements which facilitates spread of these genes between bacteria of same or different species (Coque TM, et al., 2002). These big plasmids also carry genes for resistance to other antibiotics such as aminoglycosides, trimethoprim, sulphonamides, tetracyclines and chloramphenicol (Paterson DL, 2000) which makes treatment of bacterial infections even more difficult. This has been illustrated by recent community surveys from Canada (Pitout JD, et al., 2004), Italy (Brigante G, et al., 2005), Spain (Rodríguez-Bano J, et al., 2004), Greece (Pournaras S, et al., 2004) and United Kingdom (UK) (Woodford N, et al., 2004). Recent studies in Uganda have pointed out inappropriate use of antimicrobial agents in animal industry as one of the major drivers to the emergence of Antimicrobial Resistance among microbes (Disassa N, et al., 2017).
An agro-pastoral community is one that is involved in practice of agriculture that involves both growing of crops and rearing of animals and such a community is appropriate for study of ESBL producing microorganisms since AMR is primarily as a result of continuous exposure of bacterial strains to beta-lactam antibiotics leading to production of these enzymes (Pitout JD and Laupland KB, 2008) and these communities are involved in such activities where they are more likely to use different antimicrobial agents compared to other communities.
The increased application of antimicrobial agents in treatment of human bacterial infections, agriculture and environment has primarily contributed to this phenomenon (Mukonzo JK, et al., 2013). More so, over 40% of health seeking individuals at health care facilities in Uganda are prescribed with antibiotics (Theuretzbacher U, et al., 2017). These antimicrobial agents are easily accessible even without prescription over the counter in drug shops and community pharmacies sometimes in over or under doses (Stanley IJ, et al., 2018). ESBL producing microorganisms have achieved significant attention in health care facilities and a number of studies have been done in such settings due to a number of nosocomial infections that they cause (Yadav RR and Chauhan PB, 2016) and less is known as far as community acquired infections are concerned.
Recent studies about ESBL producing Enterobacteriaceae in out-patients suggest that significant reservoirs of these microorganisms exist outside of hospitals and these infections may be an emerging challenge in community settings in various parts of the world (Rodríguez-Bano J, et al., 2004; Arpin C, et al., 2003; Colodner R, et al., 2004; Borer A, et al., 2002; Valverde A, et al., 2004). Possible community acquired ESBL producing isolate was first reported in 1998 from Ireland much as the type of the enzyme was not specified (Cormican M, et al., 1998). In addition, current research shows that these pathogens should not be exclusively considered to be involved in only nosocomial infections as they were isolated in urinary tract infections (Colodner R, et al., 2004) and bacteremia (Borer A, et al., 2002) in non-hospitalized patients. Recent findings from Spain show that 3.7%- 5.5% of the isolates from stool samples of out-patients (Valverde A, et al., 2004) were ESBL producing. Another study conducted in Canada about emergence of ESBL producing Enterobacteriaceae in community showed a prevalence of 13.7% of the ESBL phenotype (Ben-Ami R, et al., 2006).
The significance of community acquired ESBL producing Enterobacteriaceae in the epidemiology of ESBL in healthcare facilities in agro-pastoral communities of Mbarara District, Western Uganda is currently unknown. Failure to recognize the emergence of ESBL producing Enterobacteriaceae in the community and the influx of ESBL producing organisms into hospitals could undermine infection control measures in hospitals and render empirical antibiotic therapy inadequate. Therefore this study seeks to quantify and characterize ESBL producing Enterobacteriaceae in selected health facilities in agro-pastoral communities of Mbarara District, South Western Uganda.
Methodology
Study design
This was a descriptive cross-sectional study in which quantification, antimicrobial susceptibility testing and molecular characterization of Extended Spectrum Beta-lactamase were performed on Enterobacteriaceae isolates from out-patients attending Rubaya Health Centre III (HCIII), Bwizibwera Health Centre IV (HCIV) and Mbarara Regional Referral Hospital in agro-pastoral communities of Mbarara District, Western Uganda. The study included all Enterobacteriaceae isolated from urine of outpatients who were from agro-pastoral communities in Mbarara district, suspected to have urinary tract infections, and attending the study clinics. Only isolates from urine of participants who had not visited any health facility in the previous one month were considered.
Sample collection and handling
Urine samples were collected in sterile urine containers. 1 μL of urine was cultured on sterile CLED (Cysteine-Lactose-Electrolyte-Deficient) agar, incubated aerobically at 37°C overnight. The plates were checked for growth on the next day.
Identification of Enterobacteriaceae
The bacterial isolates were purified by streaking onto MacConkey agar and incubated for 18-24 hours at 35°C-37°C in aerobic conditions. Gram staining was done, lactose and non-lactose fermenting isolates from the streaked area were processed for biochemical testing.
Biochemical tests included Indole test, urease production, citrate utilization, motility, Lysine decarboxylation and carbohydrate utilization tests. Analytical Profile Index (API) system was used to confirm the identity of Enterobacteriaceae.
Phenotypic Screening of Enterobacteriaceae for Extended-Spectrum β-Lactamases (ESBLs) production
Phenotypic screening of Enterobacteriaceae for ESBL production was done using Cefotaxime (CTX) (30 μg) and Ceftazidime (CAZ) (30 μg) antibiotic discs. The isolates were cultured on Muller Hinton agar at 35℃-37℃ overnight. A sterile cotton swab was used to pick the isolates from Muller Hinton agar into a tube containing sterile normal saline (0.85% NaCl) to prepare the bacterial suspension whose turbidity shall be adjusted to 0.5 McFarland standards. Adjustng the turbidity to 0.5 McFarland was done using a densimeter machine.
The suspension was inoculated onto Muller Hinton agar plate using a sterile cotton swab by surface spreading method. The above antibiotic discs were placed on Muller Hinton agar at a distance of 30 mm apart. The plates containing the organisms and antibiotic discs were incubated at temperature of 35℃-37℃ overnight. The zone diameters were read and converted into millimeters and an interpretation of either Sensitive (S) or Resistant (R) were made. The interpretation was made using CLSI guidelines 30th version. Enterobacteriaceae, which showed resistance to any one of the used antibiotics, was considered as screen positive. These were further confirmed for ESBL production using Double Disc Synergy Test (DDST) method.
Confirmation of ESBL producing Enterobacteriaceae
Double Disc Synergy method: The turbidity of Enterobacteriaceae suspension was adjusted to 0.5 McFarland standards and was spread on Mueller Hinton agar plate using sterile cotton swab. An antibiotic disc of Ceftazidime (CAZ) (30 μg) alone and a disk of Ceftazidime in combination with Clavulanic acid (CAL) (30/10 μg) or a disc of Cefotaxime (CTX) (30 μg) alone and a disc of Cefotaxime in combination with Clavulanic acid (CTC) (30/10 μg) were used. The discs were placed about 25 mm apart and the plates containing the test organism and the discs were incubated at 35°C-37°C overnight. The zones of inhibition for both the single and combined discs were measured. An increase in zone diameter of ≥ 5 mm of the disc containing Clavulanic acid as compared to that without Clavulanic acid were considered ESBL positive (Moses A, et al., 2014). The Figure 1above represents Double Disc Synergy method for confirmation of ESBL positive Enterobacteriaceae where CAZ represents single Ceftazidime disc while CAL represents combined disc of Ceftazidime and Clavulanic acid.

Figure 1: Confirmation of ESBL producing Enterobacteriaceae using Double Disc Synergy Test (DDST)
Antibiotic susceptibility testing
The antibiotic susceptibility testing was performed on Enterobacteriaceae isolates that were ESBLs positive according to Clinical and Laboratory Standards Institute (Moses A, et al., 2014). This was done by preparing the bacterial suspension with turbidity equivalent to 0.5 Mac Farland standard. The bacterial suspension was spread on freshly prepared Muller Hinton agar using a sterile cotton swab. The ESBLs producing Enterobacteriaceae were tested on the following antibiotics: Ampicillin (10 μg), Ceftriaxone (30 μg), Cefepime (30 μg), Imipenem (10 μg), Meropenem (10 μg), Ami kacin (30 μg), Cefoxitin (30 μg), Nalidixic acid (30 μg), Ciprofloxacin (5 μg), Nitrofurantoin (300 μg), Amoxicillin-Clavulanate (20/10 μg), Piperacillin-Tazobactam (100/10 μg) Gentamicin (10 μg) and Aztreonam (30 μg). Muller Hinton agar plates containing the test organism and antibiotic discs were incubated overnight at 35°C-37°C. Zones of inhibition were measured and an interpretation of whether Sensitive (S) or Resistant (R) was done according to Clinical and Laboratory Standards Institute (Moses A, et al., 2014).
Reference strains were used for quality control of antibiotic discs used and to check the performance of culture media. The reference strains used included Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603.
Detection of genes associated with ESBLs production
Molecular characterization of ESBLs producing Enterobacteriaceae was done using Polymerase Chain Reaction (PCR). Genotyping of ESBLs producing Enterobacteriaceae was done by performing Polymerase Chain Reaction (PCR) while using primers that are specific for the detection of blaTEM, blaSHV, and blaCTX-MU genes. DNA extraction was done using boiling lysis method.
This was carried out at Medical and Molecular Laboratories, Makerere University, Uganda. The genes were amplified using primers and conditions similar to those described in the study carried out in Southwestern Uganda (Moses A, et al., 2014). The frozen Enterobacteriaceae isolates were thawed, sub-cultured and incubated at 35℃-37℃ for 18 hrs. Visualization of results was carried out using 1.2% agarose electrophoresis and bioimager.
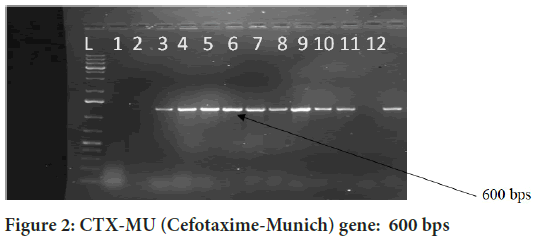
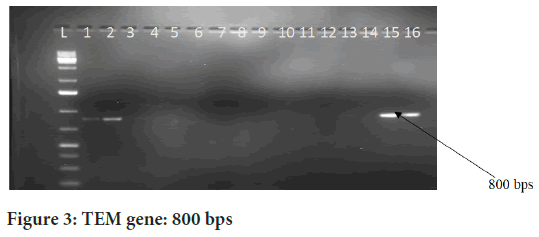
The Figures 2-4above represent gel images of ESBL genes that encode ESBL enzymes. Well L in the figure represents the Ladder; the wells 1 and 2 represent positive and negative controls respectively while wells 3-16 are sample wells. The genes CTX-MU, TEM and SHV are responsible for production of ESBL enzymes that enable the bacteria to confer resistance to third and fourth generation cephalosporins.
Figure 2: CTX-MU (Cefotaxime-Munich) gene: 600 bps
Figure 3: TEM gene: 800 bps
Figure 4: SHV (Sulfhydryl Reagent Variable) gene: 100 bps
Data analysis
Data was entered into Microsoft Excel, double-checked for completeness, cleaned and exported to STATA (v13) for analysis. Data handled included ESBL status, antibiotic susceptibility patterns and results and socio-demographic data on age, gender and agricultural background which were analyzed as follows; Descriptive statistics of demographic variables on age, gender and agricultural background were summarized and presented as frequencies and percentages. ESBL status was determined using descriptive statistics as either positive or negative and summarized as percentages. Antibiotic susceptibility patterns of ESBL positive Enterobacteriaceae were determined in terms of percentages of Sensitive or Resistant. Descriptive statistics of Genes present in the ESBL positive Enterobacteriaceae were summarized and presented as frequencies or percentages.
Age of the participant where the bacteria was isolated was important to identify the age group at risk of infection with ESBL producing Enterobacteriaceae, sex of the participant was important to know the gender mostly affected by ESBL producing Enterobacteriaceae while agricultural background was used to establish the likelihood of acquiring ESBL producing Enterobacteriaceae from livestock. All this information was extracted from Holistic Approach to Unravel Antibacterial Resistance in East Africa (HATUA) study database.
Results
Socio-demographic characteristics of participants colonized by Enterobacteriaceae
Table 1 above shows Socio-demographic characteristics of participants colonized by Enterobacteriaceae. Out of 88 participants colonized by Enterobacteriaceae, 76(86.36%) were females, the age category of 15-44 years had the highest number of participants at 60(68.18%) while 14 years and below category had the least number at 2(2.27%) colonized by Enterobacteriaceae. The number of participants who possessed livestock were 62(70.45%), Mbarara Regional Referral Hospital yielded the highest number of participants colonized by Enterobacteriaceae at 41(46.59%) followed by Bwizibwera HCIV at 33(37.50%) and Rubaya HCIII at 14(15.9%).
| Variable | Category | Frequency (n) | Proportion (%) |
|---|---|---|---|
| Gender | Female | 76 | 86.36 |
| Male | 12 | 13.64 | |
| Age | 14 years and below | 2 | 2.27 |
| 15-29 | 33 | 37.5 | |
| 30-44 | 27 | 30.68 | |
| 45-59 | 22 | 25 | |
| 60-74 | 4 | 4.55 | |
| Livestock possession | Yes | 62 | 70.45 |
| No | 26 | 29.55 | |
| Health facility | Mbarara Regional Referral Hospital | 41 | 46.59 |
| Bwizibwera Health Centre IV | 33 | 37.5 | |
| Rubaya Health Centre III | 14 | 15.9 |
Table 1: Socio-demographic characteristics of study participants colonized with Enterobacteriacea
Bacterial isolation trends from participants with community acquired Urinary Tract Infections
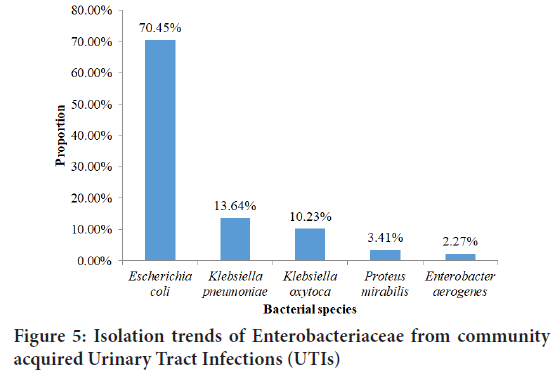
Figure 5above shows the proportion of Enterobacteriaceae isolated from community acquired urinary tract infections. Of 88 Enterobacteriaceae isolated, 62(70.45%) were Escherichia coli, 12(13.64%) were Klebsiella pneumoniae, Klebsiella oxytoca were 9(10.23%), 3(3.41%) were Proteus mirabilis while 2(2.27%) were Enterobacter aerogenes. Escherichia coli are the most prevalent uropathogen followed by Klebsiella pneumonia because they are more adapted to the tropical climate compared to other Entero bacteriaceae. Besides E. coli can easily gain entry into the urinary tract via stool.
Figure 5: Isolation trends of Enterobacteriaceae from community acquired Urinary Tract Infections (UTIs)
Prevalence of Extended Spectrum Beta-Lactamase (ESBLs) producing Enterobacteriaceae
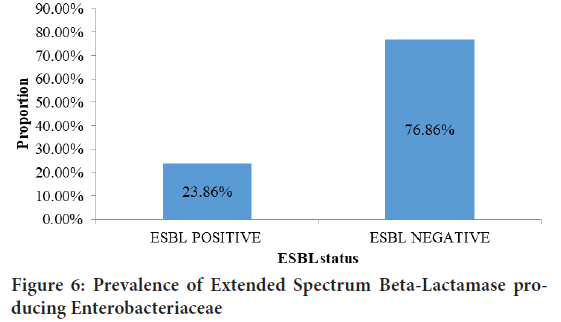
Figure 6above shows the proportion of ESBL producing Enterobacteriaceae isolated from community acquired urinary tract infections. Out of 88 Enterobacteriaceae isolated, 21(23.86%) were phenotypically confirmed as producers of Extended Spectrum Beta-Lactamases. This was confirmed by double disc synergy method.
Figure 6: Prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae
Risk factors to Extended Spectrum Beta-Lactamase (ESBLs) production
Table 2above shows risk factors associated with colonization with ESBL producing Enterobacteriaceae. There was a significant association between Extended Spectrum Beta-Lactamase production among Enterobacteriaceae and gender of the participants (P-value=0.002). Age categories, livestock possession and health facility were not associated with ESBL production among Enterobacteriaceae.
| Variable | Category | Frequency (n) | P-value |
|---|---|---|---|
| Gender | Female | 15 | 0.002 |
| Male | 6 | ||
| Age category | 14 years and below | 1 | 0.68 |
| 15-29 | 11 | ||
| 30-44 | 7 | ||
| 45-59 | 2 | ||
| 60-74 | 0 | ||
| Livestock possession | Yes | 14 | 0.663 |
| No | 7 | ||
| Health facility | Mbarara Regional Referral Hospital | 11 | 0.62 |
| Bwizibwera Health Centre IV | 6 | ||
| Rubaya Health Centre III | 4 |
Table 2: Factors associated with Extended Spectrum Beta-Lactamases (ESBLs) production
Antibiotic susceptibility patterns of Extended Spectrum Beta-Lactamase (ESBLs) producing Enterobacteriaceae
Table 3above shows antibiotic susceptibility patterns of confirmed ESBL producing Enterobacteriaceae. Extended Spectrum Beta-Lactamase producing Enterobacteriaceae were tested on 13 antibiotics to determine disc susceptibility profiles. Extended Spectrum Beta-Lactamase producing Enterobacteriaceae were most Resistant to Ampicillin, Cefepime, Aztreonam, Nalidixic acid, Ciprofloxacin and Amoxicillin/clavulanate at 100%, 100%, 95.24%, 90.48%, 85.71%, 80.95% and 61.90% respectively.
| Antibiotics | Sensitive | Resistant |
|---|---|---|
| Amoxicillin/Clavulanate (20/10 µg) | 4 (19.05%) | 17 (80.95%) |
| Ampicillin (10 µg) | 0 | 21(100%) |
| Cefepime (30 µg) | 0 | 21(100%) |
| Nalidixic acid (30 µg) | 2(9.52%) | 19(90.48%) |
| Aztreonam (10 µg) | 1(4.76%) | 20(95.24%) |
| Ciprofloxacin (5 µg) | 3(14.29) | 18(85.71%) |
| Meropenem (10 µg) | 20 (95.24%) | 1 (4.76%) |
| Piperacillin/Tazobactam (100/10 μg) | 17(80.95%) | 4(19.05%) |
| Nitrofurantoin (300 μg) | 14 (66.67%) | 7 (33.33%) |
| Gentamicin (10 µg) | 19 (90.48%) | 2 (9.52%) |
| Cefoxitin (30 μg) | 16 (76.19%) | 5(23.81%) |
| Amikacin (30 μg) | 20 (95.24%) | 1(4.76%) |
| Imipenem (10 μg) | 20(95.24%) | 1(4.76%) |
Table 3: Antibiotic susceptibility patterns of phenotypically confirmed Extended Spectrum Beta-Lactamase producing Enterobacteriaceae (N=21)
Extended Spectrum Beta-Lactamase producing Enterobacteriaceae were most sensitive to Meropenem, Imipenem, Amikacin, Gentamycin, Cefoxitin, Nitrofurantoin and Piperacillin/tazobactam at 95.24%, 95.24%, 95.24%, 90.48% 79.19%, 66.67% and 80.95% respectively. Multi Drug Resistance was observed at 85.71%.
Genotyping of Extended Spectrum Beta-producing Enterobacteriaceae
Table 4above shows the prevalence of ESBL genes. The genes responsible for ESBLs production were present in 19(90.48%) of the organisms while 2(9.52%) did not harbor the genes for ESBL even when they were phenotypically positive for ESBL production. The genes CTX-M, SHV and TEM were present in 2(9.52%) of the isolates, 3(14.29%) contained both CTX-M and SHV, 3(14.29%) contained both CTX-M and TEM, 1(4.76%) contained TEM and SHV. The organisms that contained CTX-M alone were 6(28.57), TEM alone were 3(14.29%) while 1 (4.76%) contained only SHV. All in all, 46.7% of genotypically positive ESBL Enterobacteriaceae contained CTX-M, 30% contained TEM and 23.3% contained SHV genes.
| Genes | Isolates | Proportion (%) |
|---|---|---|
| CTX-M, SHV, TEM | 2 | 9.52 |
| CTX-M, SHV | 3 | 14.29 |
| TEM, SHV | 1 | 4.76 |
| CTX-M, TEM | 3 | 14.29 |
| Cefotaxime-Munich (CTX-M) alone | 6 | 28.57 |
| TEM alone | 3 | 14.29 |
| Sulfhydryl Reagent Variable (SHV) alone | 1 | 4.76 |
| No genes | 2 | 9.52 |
| Total | 21 | 100 |
Table 4: Prevalence of ESBLs Genes (N=21)
Discussion
Socio-demographic characteristics of study participants colonized by Enterobacteriaceae
In the current study, females were the more infected with Enterobacteriaceae at 86.36% implicated in Urinary Tract Infections. This can be attributed to physiological and anatomical differences (Malik N, et al., 2015) compared to males. In addition males have poor health seeking behaviour compared to female counterparts and as such they are less likely to visit health facilities even when they are sick. This percentage is higher compared to 58% previously reported in Sri Lanka (Priyadharshana U, et al., 2019) and another related study conducted in Spain which reported bacterial isolation rates of 57% among females.
A related study conducted in Pakistan revealed a lower prevalence of Urinary Tract Infections among females at 72% (Barguigua A, et al., 2013). The most affected age category was 15-29 followed by 30-44 years at 37.50% and 30.68% respectively. This can be attributed to the fact that it is the most sexually active age hence higher chances of contracting Urinary Tract Infections (Barguigua A, et al., 2013). However the association between age and infection was not statistically significant in the current study.
It was reported that most participants infected with Enterobacteriaceae possessed livestock at 70.45%. This is because Mbarara is found in south-western region of Uganda where most occupants are agro-pastoralists.
Mbarara Regional Referral Hospital (MRRH) yielded the highest number of participants infected with Enterobacteriaceae at 46.59% followed by Bwizibwera HCIV and then Rubaya HCIII. This is because MRRH is a high volume facility compared to Bwizibwera HCIV and Rubaya HCIII and the fact that it is a regional referral hospital makes I attract more patients seeking for quality services in addition to having complicated cases.
Bacterial isolation trends
The most isolated organism from urine was Escherichia coli at 70.45% followed by Klebsiella spp, proteus mirabilis and Enterobacter aerogenes. The findings are consistent with other studies indicating that Escherichia coliis the commonest bacteria implicated in community acquired UTIs (Abraham S, et al., 2012) and other bacterial infections (Cantón R, et al., 2012; Nepal K, et al., 2017). However the isolation rate of Escherichia coli from this study was slightly lower compared to the common isolation rates which are reported between 75%-90% (Chakrawarti A, et al., 2015).
A related study in Uganda demonstrated that Escherichia coli was the most isolated organism among outpatients at 89% which is higher than the isolation rate reported in our study (Najjuka CF, et al., 2016). It has been reported previously that Escherichia coli and Klebsiella spp are the major ESBL producing Enterobacteriaceae isolated from both community and hospital settings (Koksal E, et al., 2019; Kuster SP, et al., 2010; Hyle EP, et al., 2005; Pitout JD, et al., 2004) which is in agreement with results from this study. A related study in Pakistan demonstrated 33.5% prevalence of ESBL among Escherichia coli and 15.25% among Klebsiella spp.
Proportion of Extended Spectrum Beta-Lactamase (ESBLs) producing Enterobacteriaceae
The current study reported the prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae at 23.86% demonstrating an increase in community spread of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae in Uganda as compared to a previous study by Najjuka CF, et al., 2016.
The results from this study demonstrated higher prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae from the community compared to 8.5% prevalence reported in Nigeria (Nwafia IN, et al., 2019).
A related study in Morocco demonstrated a lower prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae at 7.5% (Barguigua A, et al., 2013) compared to our study. This can be attributed to difference in climatic conditions, socio-economic factors and regulations regarding antibiotic use which act as major sources of selection pressure for production of ESBLs.
Related studies have reported higher prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae compared to this study.
For instance a study carried out in Iran demonstrated 52% prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae from community acquired Urinary Tract Infections (Latifpour M, et al., 2016).
A related study in Canada demonstrated prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae from the community at 71% (Pitout JD, et al., 2004) while another study in Nepal reported 35.9% prevalence (Nepal K, et al., 2017).
A related study in Nigeria demonstrated prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae from community settings at 35% (Afunwa RA, et al., 2011). Related studied in India and Turkey demonstrated higher prevalence compared to this study at 40% (Balasubramanian S, et al., 2018) and 37.1% (Koksal E, et al., 2019) respectively. The differences in prevalence can be attributed to differences in climatic conditions, health seeking behaviour, socio-economic factors, antibiotic availability and use which either singly or in combination contributes to selection pressure for ESBLs production. In addition, countries with highest antibiotic consumption rates have been found to have high prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae for instance Turkey, Tunisia, Algeria, Greece and Romania (Klein EY, et al., 2018; ECDC, 2017; Albiger B, et al., 2015).
It has also been noted that the spread of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae is attributed to the existence of reservoirs in the community especially by recently discharged patients (Dziri R, et al., 2016). Other potential reservoirs have been identified for instance a study in Tanzania demonstrated existence of ESBL producing Enterobacteriaceae in public latrines (Erb S, et al., 2018). Other studies have demonstrated faecal carriage of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae among humans which potentiates the spread of such organisms (Valverde A, et al., 2004; Ben-Ami R, et al., 2006; Hazirolan GÜ, et al., 2018; de Champs C, et al., 1989; Holländer R, et al., 2001).
In addition it has been of great concern that ESBL producing Enterobacteriaceae are transmitted from animals to humans leading to cross infestation (Koovapra S, et al., 2016; Shrivastav A, et al., 2016; Bogaerts P, et al., 2015; Hordijk J, et al., 2013).
In addition, Extended Spectrum Beta-Lactamase producing Enterobacteriaceae have recently been isolated in food products which is a potential source to humans especially when these products are consumed in raw form (Leverstein-van MAH, et al., 2011; Reuland EA, et al., 2014; Kluytmans JA, et al., 2013; Evers EG, et al., 2017; Stanley IJ, et al., 2018).
Risk factors to acquisition of infection by Extended Spectrum Beta-Lactamase (ESBLs) producing Enterobacteriaceae
Among the risk factors assessed, female gender was associated to increased risk of infection by Extended Spectrum Beta-Lactamase producing Enterobacteriaceae with P-value<0.005. This observation is similar to a study conducted in Canada which demonstrated that female gender was associated with infection by Extended Spectrum Beta-Lactamase producing Enterobacteriaceae (Brolund A, 2014). Other related studies have demonstrated an association between female gender with infection with Extended Spectrum Beta-Lactamase producing Enterobacteriaceae (BenAmi R, et al., 2009; Yilmaz E, et al., 2008; Colodner R, et al., 2004).
Contrary to this study, related studies in Uganda (Najjuka CF, et al., 2016) and India (Balasubramanian S, et al., 2018) demonstrated that there was no single risk factor associated with Extended Spectrum Beta-Lactamase production by Enterobacteriaceae.
Other factors assessed were not statistically significant to infection with Extended Spectrum Beta-Lactamase producing Enterobacteriaceae which included age category, livestock possession and health facility with P-values 0.680, 0.663, 0.620 respectively.
Other related studies worldwide have reported a number of factors related to infection by Extended Spectrum Beta-Lactamase producing Enterobacteriaceae for instance a study in United States of America demonstrated that prior exposure to broad spectrum antibiotics, recent hospitalization and surgery were potential risk factors to infection with Extended Spectrum Beta-Lactamase Producing Enterobacteriaceae (Biehl LM, et al., 2016).
A similar study conducted in Nigeria demonstrated that recent surgery, hospitalization, recent antibiotic uses were associated with colonization and infection with Extended Spectrum Beta-Lactamase producing Enterobacteriaceae (Nwafia IN, et al., 2019). A related study in Australia reported that recent fluoroquinolone use was related to production of ESBLs among Enterobacteriaceae due to accidental expression of genes because ESBL genes are located on the same plasmid with those genes responsible for quinolone resistance (Paterson DL, 2000) and a related study demonstrated that ESBL production was directly related to fluoroquinolone use (Babini GS and Livermore DM, 2000).
Antibiotic susceptibility patterns of Extended Spectrum Beta-Lactamase (ESBLs) producing Enterobacteriaceae
A total of 13 antibiotics selected from 7 different classes were tested on ESBL producing Enterobacteriaceae. Extended Spectrum Beta-Lactamase producing Enterobacteriaceae were most Resistant to Ampicillin, Cefepime, Aztreonam, Nalidixic acid, Ciprofloxacin and Amoxicillin/clavulanic acid. Several studies worldwide have demonstrated comparable findings. For instance a review in United States of America pointed out that fluoroquinolones were no longer consistently reliable in treatment of bacterial infections (Bonomo RA, et al., 2017).
A related study conducted in Nigeria demonstrated consistent results to this study where it was shown that Extended Spectrum Beta-Lactamase producing Enterobacteriaceae were Resistant to quinolones, penicillins and beta-lactam combinations (Hoban DJ, et al., 2012; Priyadharshana U, et al., 2019; Pitout JD, et al., 2004; Afunwa RA, et al., 2011). A related study in India demonstrated high resistance rates to flouroquinolones and beta-lactam agents (Balasubramanian S, et al., 2018). A related study in Japan demonstrated similar findings to this study where it was revealed that Cefepime was ineffective at treatment of infections caused by ESBL producing Enterobacteriaceae (Matsumura Y, et al., 2015).
The findings from this study differ from other studies done worldwide for instance studies which have recommended the use of Cefepime to treat complicated bacterial infections caused by ESBL producing Enterobacteriaceae (Kanj SS and Kanafani ZA, 2011; Nguyen HM, et al., 2014; Harris PN, et al., 2018; Vardakas KZ, et al., 2012).
This study revealed high sensitivity rates of ESBL producing Enterobacteriaceae to Imipenem, Meropenem, Amikacin, Gentamycin, Piperacillin/ tazobactam, Cefoxitin and Nitrofurantoin. The findings from this study are in agreement with several studies conducted worldwide for instance a study conducted in Nepal revealed that all isolated tested were Sensitive to Imipenem (Nepal K, et al., 2017) and a similar study conducted in Nigeria demonstrated 97.14% sensitivity to Imipenem (Nwafia IN, et al., 2019) and thus carbapenems have been recommended as drug of choice to treat infections caused by ESBL producing Enterobacteriaceae (Papp-Wallace KM, et al., 2019; Kalaskar A and Venkataramana K, 2012) although carbapenems have been linked to increased risk of fungal infections (Ng TM, et al., 2016). In addition there have been concerns of emergence of carbapenem resistance among Enterobacteriaceae which makes treatment of complicated bacterial infections more difficult (Brolund A, 2014; Abraham S, et al., 2012; Rodríguez-Baño J, et al., 2018).
A related study in Iran demonstrated similar findings to this study where high sensitivity rates were reported to Gentamycin (Eftekhar F, et al., 2012). A related study in India demonstrated similar findings to this study where high sensitivity rates were reported to Imipenem, Amikacin and Nitrofurantoin (Ranjini CY, et al., 2015).
Related studies have tested and recommended Beta-lactam combinations such as Piperacillin/tazobactam for the treatment of infections caused by Extended Spectrum Beta-Lactamase producing Enterobacteriaceae (Sucher AJ, et al., 2015; Scott LJ, 2016; Solomkin J, et al., 2015; Lucasti C, et al., 2014).
The findings from this study are contrary to a related study conducted in Iran which reported high resistance rates to Nitrofurantoin and amikacin (Eftekhar F, et al., 2012).
Related studies in Spain and South Korea recommended the use of Piperacillin/tazobactam as an alternative to carbapenems for the treatment of complicated infections caused by ESBL producing Enterobacteriaceae (Seo YB, et al., 2017; Ko JH, et al., 2018) although it should be used in stable conditions and where antibiotic susceptibility profiles have been carried out (Pilmis B, et al., 2017) because it has been associated to high mortality rates in some studies (Rodríguez-Bano J, et al., 2004).
Multi-Drug Resistance (MDR) among ESBL producing Enterobacteriaceae from this study was very high and established at 85.71%. This is in agreement to other studies conducted worldwide for instance a study carried out in Morocco established Multi-Drug Resistance among ESBL producing Enterobacteriaceae at 91.1% (Barguigua A, et al., 2013) while a related study in India established Multi-Drug Resistance at 82.6% (Ranjini CY, et al., 2015). These observations are consistent to the previously reported co-resistance to other antibiotics (Calbo E, et al., 2006). This has been attributed to the fact that genes responsible for ESBL production are encoded on big plasmids which contain other resistance genes to other antibiotics (Kalaskar A and Venkataramana K, 2012).
The high resistance rates observed in this study can be attributed to overuse and misuse of antibiotics to treat common infections (Cars O, et al., 2008). It can also be attributed to socio-economic differences where patients lack consultation fees on which treatment to consider in addition to prior knowledge of which antibiotics to use hence resorting to self-medication (Grigoryan L, et al., 2008). Thus it has been reported that countries with the most reserved antibiotic prescribing patterns have relatively low rates of resistance (Goossens H, et al., 2005).
Genotyping of Extended Spectrum Beta-Lactamase (ESBLs) producing Enterobacteriaceae
The genes responsible for production of Extended Spectrum Beta-Lactamases among Enterobecteriaceae were determined using Polymerase Chain Reaction (PCR) where 90.48% of the isolates harboured the genes CTM-M, TEM and SHV which is in agreement with what has been reported from other studies that these genes are the most prevalent ESBL genes in both community and hospital settings (Rodríguez-Bano J, et al., 2004).
This study demonstrated that an organism can harbour more than one ESBL gene which is consistent with other studies carried out previously for instance a related study carried out in Western Uganda demonstrated that 40.8% of the organisms harboured more than one gene (Moses A, et al., 2014). A related study in Morocco also demonstrated that an organism can harbour more than ESBL gene (Barguigua A, et al., 2013).
The gene CTX-M was the most prevalent at 46.7%, followed by TEM at 30% and SHV at 23.3%. The findings from this study are consistent with other studies conducted worldwide. For instance a study conducted in Libya demonstrated that CTX-M was the commonest isolate (Ahmed OB, et al., 2013) and similarly higher rates of CTX-M have been found in Europe (Valenza G, et al., 2014; Rodríguez-Bano J, et al., 2004), and in other parts of Africa (Lonchel CM, et al., 2013; Ahmed OB, et al., 2013). The high prevalence of CTX-M in this study can be attributed to the widespread use of ceftriaxone to treat serious bacterial infections (Bonnet R, 2004).
The prevalence of TEM and SHV was found to be lower than that of CTX-M because TEM and SHV are more prevalent in hospital settings compared to community settings (Wiener J, et al., 1999). Different rates of SHV and TEM have been reported worldwide for instance a study conducted in Sri Lanka indicated higher prevalence of TEM compared to SHV (Priyadharshana U, et al., 2019) and a related study conducted in Western Uganda demonstrated higher prevalence of TEM compared to SHV (Moses A, et al., 2014). Some studies have reported equal prevalence of TEM and SHV for instance a related study in Spain demonstrated 18% proportions for both SHV and TEM genes (Rodríguez-Bano J, et al., 2004). A related study conducted in Iran demonstrated contrary findings to this study where higher prevalence of SHV compared to TEM were reported (Latifpour M, et al., 2016).
This study also demonstrated that ESBL genes in community settings are similar to those in hospital care settings irrespective of the proportions which suggests possible importation of ESBL producing Enterobacteriaceae from the community into the hospital and vice versa for instance a related study in Israel involved screening of patients for ESBL at admission and it was discovered that 10.8% of the patients were colonized by ESBL producing Enterobacteriaceae (Ben-Ami R, et al., 2006).
The findings from this study clarified that there are other types of ESBL genes that were not detected using the primers specific for CTX-M, TEM and SHV hence a discrepancy between phenotypic and genotypic methods for ESBL detection.
It was demonstrated that 2(9.52%) Enterobacteriaceae did not harbour any of the 3 genes under detection. This can be attributed to the fact that new ESBL genes are emerging as a result of mutations. The findings from this study are consistent with other studies conducted worldwide for instance a study conducted in Iran demonstrated that 8% of the organisms that were ESBL positive at phenotypic detection contained Vietnamese Extended-Spectrum β-Lactamase (VEB) gene which would have been missed out if only 3 primers for CTX-M, TEM and SHV were used (Latifpour
M, et al., 2016).
A related study in Israel demonstrated phenotypically ESBL positive isolates that lacked common ESBL genes at genotyping (Ben-Ami R, et al., 2006). Similarly, a related study conducted in Sudan indicated that 47.7% of phenotypically ESBL positive isolates did not harbour common ESBL genes (Ahmed OB, et al., 2013). A related study in India demonstrated that not all phenotypically positive ESBL isolates harbour the three common genes as 61.6% of the isolates did not contain CTX-M, TEM or SHV genes (Shahid M, et al., 2011). Several other studies have demonstrated that there are other ESBL genes that need to be detected as far as genotyping for ESBL is concerned (Eftekhar F, et al., 2012; Moses A, et al., 2014).
This study demonstrated that all genotypically positive Enterobacteriaceae were phenotypically positive which is contrary to other studies for instance a study carried out in Western Uganda reported that 37% of the isolates contained ESBL genes but were negative on phenotypic assessment (Moses A, et al., 2014). This can be attributed to lack of effective selection pressure to trigger gene expression (Kalaskar A and Venkataramana K, 2012).
The high prevalence of ESBL genes in community settings can be attributed to the fact that there is horizontal transfer of ESBL genes between Enterobacteriaceae existing in the environment. This is because ESBL genes are located on mobile genetic elements which favour their dissemination between Enterobacteriaceae. For instance a related study in Turkey demonstrated that ESBL genes were present in Enterobacteriaceae isolated from faecal samples and this potentiates spread of genes to other isolates colonizing humans (Hazirolan GÜ, et al., 2018).
A related study in Korea demonstrated the potential spread of ESBL producing Enterobacteriaceae and exchange of ESBL genes from farm animals and farm environment to humans (Tamang MD, et al., 2013). Since this study was carried out in an agro-pastoral community, such spread cannot be ruled out.
Conclusion
The prevalence of Extended Spectrum Beta-Lactamase producing Enterobacteriaceae from our study is high while the most isolated bacterial species are Escherichia coli and Klebsiella spp. Carbapenems are effective against Extended Spectrum Beta-Lactamase producing Enterobacteriaceae while Beta Lactam/Beta Lactamase inhibitor combination antibiotics are second in effectiveness to Carbapenems. The rate of Multi Drug Resistance (MDR) to common antibiotics used to treat Urinary Tract Infections is high. The most prevalent genes responsible for ESBL production are CTX-M and TEM.
Recommendations
Double disc synergy method should be implemented in routine confirmation of ESBL producing Enterobacteriaceae in health facilities in resource scarce settings. This would encourage definitive therapy instead of empirical therapy in management of bacterial infections.
More studies about antimicrobial resistance in community settings should be carried out. This will help to establish action areas and points where to reduce spread of resistance either from the community to the healthcare settings.
Infection control points should be established especially at the entrance of health facilities to reduce influx of ESBL producing Enterobacteriaceae from the community. Likewise personal hygiene like hand washing, cleanliness should be emphasized in health facilities and community to reduce spread of ESBL producing Enterobacteriaceae.
Antimicrobial surveillance should be done both in both community and health care settings to be able to identify action areas to combat spread of antimicrobial resistance. Data obtained should direct policy makers for effective decision making.
The antibiotics that were identified as ineffective for instance Ampicillin, Amoxicillin/clavulanic acid, Cefepime, Nalidixic acid, Ciprofloxacin and Aztreonam should be rested in order to reduce the selection pressure against them by the bacteria. In future, these can be reintroduced as guided by data when they have regained their potency. Piperacillin/tazobactam should be emphasized for treatment of infections caused by ESBL producing Enterobacteriaceae as an alternative to carbapenems to reduce the risk of development of carbapenem resistance among Enterobacteriaceae.
Study Limitations
The study was limited to three health facilities in Mbarara district. This may not reflect the real picture in all facilities in south western Uganda which makes generalization of findings difficult. Only Enterobacteriaceae isolated from outpatients were considered hence the results cannot be generalized to hospitalized patients. Data about history of recent antibiotic use was not collected hence making it difficult to determine association as far as resistance development is considered. The study was limited to human beings and no comparative study was carried out in livestock to establish existence of ESBL.
Declarations
Acknowledgements
The authors wish to thank the participants that participated in this study and the institutions involved in the study which include St. Andrews University in Scotland, Makerere University and Mbarara University of Science and Technology in Uganda.
Funding
This work was supported by the “Holistic Approach to Unravel Antibacterial Resistance in East Africa” project which was a 3-year Global Context Consortia Award (MR/S004785/1) funded by the National Institute of Health, Medical Research Council and the Department of Health and Social Care, UK.
Availability of data and materials
All data supporting the conclusions of this article are included within the article.
Authors’ contributions
IM collected samples and data, participated in performing all the laboratory analyses and wrote the first draft of the manuscript; JB supervised the study; HZ participated in data analysis; FB critically revised the manuscript; WS participated in interpreting PFGE data; MH participated in supervision of the study and BBA supervised the study and critically revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and informed consent
The study was approved by Institutional Ethical Review Committee (IERC) of Mbarara University of Science and Technology, reference number 45/03-20. Informed consent was obtained from all the participants aged 18 and above and legal guardians for participants aged below 18. The study was conducted in accordance with the national guidelines for research involving humans as research participants and national guidelines to conduct research during COVID-19 pandemic as issued by Uganda National Council of Science and Technology (UNCST).
References
- Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013; 13(12): 1057-1098.
[Crossref] [Google Scholar] [Pubmed]
- Klein EY, Tseng KK, Pant S, Laxminarayan R. Tracking global trends in the effectiveness of antibiotic therapy using the Drug Resistance Index. BMJ Glob Health. 2019; 4(2): e001315.
[Crossref] [Google Scholar] [Pubmed]
- WHO. High levels of antibiotic resistance found worldwide, new data shows, 2018. World Health Organization. 2018.
- Ampaire L, Muhindo A, Orikiriza P, Mwanga-Amumpaire J, Boum Y, Bebell L. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016; 5(1): 1-6.
[Crossref] [Google Scholar] [Pubmed]
- Frost I, van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global geographic trends in antimicrobial resistance: The role of international travel. J Travel Med. 2019; 26(8): 36.
[Crossref] [Google Scholar] [Pubmed]
- Kateregga JN, Kantume R, Atuhaire C, Lubowa MN, Ndukui JG. Phenotypic expression and prevalence of ESBL-producing Enterobacteriaceae in samples collected from patients in various wards of Mulago Hospital, Uganda. BMC Pharmacol Toxicol. 2015; 16(1): 1-6.
[Crossref] [Google Scholar] [Pubmed]
- Najjuka CF, Kateete DP, Kajumbula HM, Joloba ML, Essack SY. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes. 2016; 9(1): 1-4.
[Crossref] [Google Scholar] [Pubmed]
- Ben-Ami R, Schwaber MJ, Navon-Venezia S, Schwartz D, Giladi M, Chmelnitsky I, et al. Influx of extended-spectrum ß-lactamase-producing Enterobacteriaceae into the hospital. Clin Infect Dis. 2006; 42(7): 925-934.
[Crossref] [Google Scholar] [Pubmed]
- Andrew B, Kagirita A, Bazira J. Prevalence of extended-spectrum beta-lactamases-producing microorganisms in patients admitted at KRRH, Southwestern Uganda. Int J Microbiol. 2017.
[Crossref] [Google Scholar] [Pubmed]
- Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci. 2015; 22(1): 90-101.
[Crossref] [Google Scholar] [Pubmed]
- Brolund A. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect Ecol Epidemiol. 2014; 4(1): 24555.
[Crossref] [Google Scholar] [Pubmed]
- Pagani L, Migliavacca R, Pallecchi L, Matti C, Giacobone E, Amicosante G, et al. Emerging extended-spectrum ß-lactamases in Proteus mirabilis. J Clin Microbiol. 2002; 40(4): 1549-1552.
[Crossref] [Google Scholar] [Pubmed]
- Coque TM, Oliver A, Pérez-Díaz JC, Baquero F, Cantón R. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum ß-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob Agents Chemother. 2002; 46(2): 500-510.
[Crossref] [Google Scholar] [Pubmed]
- Paterson DL. Recommendation for treatment of severe infections caused by Enterobacteriaceae producing Extended-Spectrum ß-Lactamases (ESBLs). Clin Microbiol Infect. 2000; 6(9): 460-463.
[Crossref] [Google Scholar] [Pubmed]
- Pitout JD, Hanson ND, Church DL, Laupland KB. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum ß-lactamases: Importance of community isolates with blaCTX-M genes. Clin Infect Dis. 2004; 38(12): 1736-1741.
[Crossref] [Google Scholar] [Pubmed]
- Brigante G, Luzzaro F, Perilli M, Lombardi G, Colì A, Rossolini GM, et al. Evolution of CTX-M-type ß-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int J Antimicrob Agents. 2005; 25(2): 157-162.
[Crossref] [Google Scholar] [Pubmed]
- Rodríguez-Bano J, Navarro MD, Romero L, Martínez-Martínez L, Muniain MA, Perea EJ, et al. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J Clin Microbiol. 2004; 42(3): 1089-1094.
[Crossref] [Google Scholar] [Pubmed]
- Pournaras S, Ikonomidis A, Sofianou D, Tsakris A, Maniatis AN. CTX-M-type ß-lactamases affect community Escherichia colitreatment, Greece. Emerg Infect Dis. 2004; 10(6): 1163.
[Crossref] [Google Scholar] [Pubmed]
- Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum ß-lactamases in the UK. J Antimicrob Chemother. 2004; 54(4): 735-743.
[Crossref] [Google Scholar] [Pubmed]
- Disassa N, Sibhat B, Mengistu S, Muktar Y, Belina D. Prevalence and antimicrobial susceptibility pattern of E. coli O157: H7 isolated from traditionally marketed raw cow milk in and around Asosa town, western Ethiopia. Vet Med Int. 2017.
[Crossref] [Google Scholar] [Pubmed]
- Pitout JD, Laupland KB. Extended-spectrum ß-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect Dis. 2008; 8(3): 159-166.
[Crossref] [Google Scholar] [Pubmed]
- Mukonzo JK, Namuwenge PM, Okure G, Mwesige B, Namusisi OK, Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. J Multidiscip Healthc. 2013; 6: 303.
[Crossref] [Google Scholar] [Pubmed]
- Theuretzbacher U, Årdal C, Harbarth S. Linking sustainable use policies to novel economic incentives to stimulate antibiotic research and development. Infect Dis Rep. 2017; 9(1): 6836.
[Crossref] [Google Scholar] [Pubmed]
- Stanley IJ, Kajumbula H, Bazira J, Kansiime C, Rwego IB, Asiimwe BB. Multidrug resistance among Escherichia coli and Klebsiella pneumoniae carried in the gut of out-patients from pastoralist communities of Kasese district, Uganda. PloS One. 2018; 13(7): e0200093.
[Crossref] [Google Scholar] [Pubmed]
- Yadav RR, Chauhan PB. The detection of Extended Spectrum Beta-Lactamases (ESBLs) producing Escherichia coli isolated from clinical samples. Int J Adv Res Biol Sci. 2016; 3(5): 9-15.
- Arpin C, Dubois V, Coulange L, André C, Fischer I, Noury P, et al. Extended-spectrum ß-lactamase-producing Enterobacteriaceae in community and private health care centers. Antimicrob Agents Chemother. 2003; 47(11): 3506-3514.
[Crossref] [Google Scholar] [Pubmed]
- Colodner R, Rock W, Chazan B, Keller N, Guy N, Sakran W, et al. Risk factors for the development of extended-spectrum beta-lactamase-producing bacteria in nonhospitalized patients. Eur J Clin Microbiol Infect Dis. 2004; 23(3): 163-167.
[Crossref] [Google Scholar] [Pubmed]
- Borer A, Gilad J, Menashe G, Peled N, Riesenberg K, Schlaeffer F. Extended-spectrum beta-lactamase-producing Enterobacteriaceae strains in community-acquired bacteremia in Southern Israel. Med Sci Monit. 2002; 8(1): CR44-CR47.
[Google Scholar] [Pubmed]
- Valverde A, Coque TM, Sánchez-Moreno MP, Rollán A, Baquero F, Cantón R. Dramatic increase in prevalence of fecal carriage of extended-spectrum ß-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J Clin Microbiol. 2004; 42(10): 4769-4775.
[Crossref] [Google Scholar] [Pubmed]
- Cormican M, Morris D, Corbett-Feeeney G, Flynn J. Extended spectrum beta-lactamase production and fluorquinolone resistance in pathogens associated with community acquired urinary tract infection. Diagn Microbiol Infect Dis. 1998; 32(4): 317-319.
[Crossref] [Google Scholar] [Pubmed]
- Moses A, Bwanga F, Boum Y, Bazira J. Prevalence and genotypic characterization of extended-Spectrum Beta-lactamases produced by gram negative bacilli at a tertiary Care Hospital in Rural South Western Uganda. Br Microbiol Res J. 2014; 4(12): 1541.
[Crossref] [Google Scholar] [Pubmed]
- Malik N, Ahmed M, ur Rehman M. Prevalence and antimicrobial susceptibility of uropathogens in patients reporting to a tertiary care facility in Peshawar, Pakistan. J Microbiol Antimicrob. 2015; 7(1): 6-12.
- Priyadharshana U, Piyasiri LB, Wijesinghe C. Prevalence, antibiotic sensitivity pattern and genetic analysis of extended spectrum beta lactamase producing Escherichia coli and Klebsiella spp among patients with community acquired urinary tract infection in Galle district, Sri Lanka. Ceylon Med J. 2019; 64(4): 140-145.
[Crossref] [Google Scholar] [Pubmed]
- Barguigua A, el Otmani F, Talmi M, Reguig A, Jamali L, Zerouali K, et al. Prevalence and genotypic analysis of plasmid-mediated ß-lactamases among urinary Klebsiella pneumoniae isolates in Moroccan community. J Antibiot. 2013; 66(1): 11-16.
[Crossref] [Google Scholar] [Pubmed]
- Abraham S, Chapman TA, Zhang R, Chin J, Mabbett AN, Totsika M, et al. Molecular characterization of Escherichia coli strains that cause symptomatic and asymptomatic urinary tract infections. J Clin Microbiol. 2012; 50(3): 1027-1030.
[Crossref] [Google Scholar] [Pubmed]
- Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: Origin and diffusion. Front Microbiol. 2012; 3: 110.
[Crossref] [Google Scholar] [Pubmed]
- Nepal K, Pant ND, Neupane B, Belbase A, Baidhya R, Shrestha RK, et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017; 16(1): 1-7.
[Crossref] [Google Scholar] [Pubmed]
- Chakrawarti A, Dongol P, Khanal H, Subba P, Benerjee JJ. Extended spectrum beta lactamases detection and multiple antibiotic resistance indexing of Escherichia coli from urine samples of patients from a referral hospital of eastern Nepal. Int J Appl Sci Biotechnol. 2015; 3(3): 423-426.
- Koksal E, Tulek N, Sonmezer MC, Temocin F, Bulut C, Hatipoglu C, et al. Investigation of risk factors for community-acquired urinary tract infections caused by extended-spectrum beta-lactamase Escherichia coli and Klebsiella species. Investig Clin Urol. 2019; 60(1): 46-53.
[Crossref] [Google Scholar] [Pubmed]
- Kuster SP, Hasse B, Huebner V, Bansal V, Zbinden R, Ruef C, et al. Risks factors for infections with extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae at a tertiary care university hospital in Switzerland. Infection. 2010; 38(1): 33-40.
[Crossref] [Google Scholar] [Pubmed]
- Hyle EP, Lipworth AD, Zaoutis TE, Nachamkin I, Bilker WB, Lautenbach E. Impact of inadequate initial antimicrobial therapy on mortality in infections due to extended-spectrum ß-lactamase-producing Enterobacteriaceae: Variability by site of infection. Arch Intern Med. 2005; 165(12): 1375-1380.
[Crossref] [Google Scholar] [Pubmed]
- Nwafia IN, Ohanu ME, Ebede SO, Ozumba UC. Molecular detection and antibiotic resistance pattern of extended-spectrum beta-lactamase producing Escherichia coli in a Tertiary Hospital in Enugu, Nigeria. Ann Clin Microbiol Antimicrob. 2019; 18(1): 1-7.
[Crossref] [Google Scholar] [Pubmed]
- Latifpour M, Gholipour A, Damavandi MS. Prevalence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates in nosocomial and community-acquired urinary tract infections. Jundishapur J Microbiol. 2016; 9(3).
[Crossref] [Google Scholar] [Pubmed]
- Afunwa RA, Odimegwu DC, Iroha RI, Esimone CO. Antimicrobial resistance status and prevalence rates of extended spectrum beta-lactamase producers isolated from a mixed human population. Bosn J Basic Med Sci. 2011; 11(2): 91.
[Crossref] [Google Scholar] [Pubmed]
- Balasubramanian S, Kuppuswamy D, Padmanabhan S, Chandramohan V, Amperayani S. Extended-spectrum beta-lactamase-producing community-acquired urinary tract infections in children: Chart review of risk factors. J Glob Infect Dis. 2018; 10(4): 222.
[Crossref] [Google Scholar] [Pubmed]
- Klein EY, van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci USA. 2018; 115(15): E3463-E3470.
[Crossref] [Google Scholar] [Pubmed]
- ECDC. Antimicrobial resistance surveillance in Europe 2015: Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). European Centre for Disease Prevention and Control. 2017.
- Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL. Carbapenemase-producing Enterobacteriaceae in Europe: Assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015; 20(45): 30062.
[Crossref] [Google Scholar] [Pubmed]
- Dziri R, Klibi N, Alonso CA, Said LB, Bellaaj R, Slama KB, et al. Characterization of Extended-Spectrum ß-Lactamase (ESBL)-producing Klebsiella, Enterobacter, and Citrobacter obtained in environmental samples of a Tunisian hospital. Diagn Microbiol Infect Dis. 2016; 86(2): 190-193.
[Crossref] [Google Scholar] [Pubmed]
- Erb S, D’Mello-Guyett L, Malebo HM, Njee RM, Matwewe F, Ensink J, et al. High prevalence of ESBL-Producing E. coli in private and shared latrines in an informal urban settlement in Dar es Salaam, Tanzania. Antimicrob Resist Infect Control. 2018; 7(1): 1-6.
[Crossref] [Google Scholar] [Pubmed]
- Hazirolan GÜ, Mumcuoglu I, Altan G, Özmen BB, Aksu N, Karahan ZC. Fecal Carriage of Extended-spectrum Beta-lactamase and AmpC Beta-lactamase-producing Enterobacteriaceae in a Turkish Community. Niger J Clin Pract. 2018; 21(1): 81-86.
[Crossref] [Google Scholar] [Pubmed]
- de Champs C, Sauvant MP, Chanal C, Sirot D, Gazuy N, Malhuret R, et al. Prospective survey of colonization and infection caused by expanded-spectrum-beta-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J Clin Microbiol. 1989; 27(12): 2887-2890.
[Crossref] [Google Scholar] [Pubmed]
- Holländer R, Ebke M, Barck H, von Pritzbuer E. Asymptomatic carriage of Klebsiella pneumoniae producing extended-spectrum b-lactamase by patientsin a neurological early rehabilitation unit: Management of an outbreak. J Hosp Infect. 2001; 48(3): 207-213.
[Crossref] [Google Scholar] [Pubmed]
- Koovapra S, Bandyopadhyay S, Das G, Bhattacharyya D, Banerjee J, Mahanti A, et al. Molecular signature of extended spectrum ß-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect Genet Evol. 2016; 44: 395-402.
[Crossref] [Google Scholar] [Pubmed]
- Shrivastav A, Sharma RK, Sahni YP, Shrivastav N, Gautam V, Jain S. Study of antimicrobial resistance due to extended spectrum beta-lactamase-producing Escherichia coli in healthy broilers of Jabalpur. Vet World. 2016; 9(11): 1259.
[Crossref] [Google Scholar] [Pubmed]
- Bogaerts P, Huang TD, Bouchahrouf W, Bauraing C, Berhin C, El Garch F, et al. Characterization of ESBL-and AmpC-producing Enterobacteriaceae from diseased companion animals in Europe. Microb Drug Resist. 2015; 21(6): 643-650.
[Crossref] [Google Scholar] [Pubmed]
- Hordijk J, Schoormans A, Kwakernaak M, Duim B, Broens E, Dierikx C, et al. High prevalence of fecal carriage of extended spectrum ß-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front Microbiol. 2013; 4: 242.
[Crossref] [Google Scholar] [Pubmed]
- Leverstein-Van MAH, Dierikx CM, Stuart JC, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011; 17(6): 873-880.
[Crossref] [Google Scholar] [Pubmed]
- Reuland EA, al Naiemi N, Raadsen SA, Savelkoul PH, Kluytmans JA, Vandenbroucke-Grauls CM. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur J Clin Microbiol Infect Dis. 2014; 33(10): 1843-1846.
[Crossref] [Google Scholar] [Pubmed]
- Kluytmans JA, Overdevest IT, Willemsen I, Kluytmans-Van MFDB, van der Zwaluw K, Heck M, et al. Extended-spectrum ß-lactamase-producing Escherichia coli from retail chicken meat and humans: Comparison of strains, plasmids, resistance genes, and virulence factors. Clin Infect Dis. 2013; 56(4): 478-487.
[Crossref] [Google Scholar] [Pubmed]
- Evers EG, Pielaat A, Smid JH, van Duijkeren E, Vennemann FB, Wijnands LM, et al. Comparative exposure assessment of ESBL-producing Escherichia coli through meat consumption. PloS One. 2017; 12(1): e0169589.
[Crossref] [Google Scholar] [Pubmed]
- Ben-Ami R, Rodríguez-Baño J, Arslan H, Pitout JD, Quentin C, Calbo ES, et al. A multinational survey of risk factors for infection with extended-spectrum ß-lactamase-producing Enterobacteriaceae in nonhospitalized patients. Clin Infect Dis. 2009; 49(5): 682-690.
[Crossref] [Google Scholar] [Pubmed]
- Yilmaz E, Akalin H, Özbey S, Kordan Y, Sinirtas M, Gürcüoglu E, et al. Risk factors in community-acquired/onset urinary tract infections due to extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. J Chemother. 2008; 20(5): 581-585.
[Crossref] [Google Scholar] [Pubmed]
- Biehl LM, Schmidt-Hieber M, Liss B, Cornely OA, Vehreschild MJ. Colonization and infection with extended spectrum beta-lactamase producing Enterobacteriaceae in high-risk patients: Review of the literature from a clinical perspective. Crit Rev Microbiol. 2016; 42(1): 1-6.
[Crossref] [Google Scholar] [Pubmed]
- Babini GS, Livermore DM. Antimicrobial resistance amongst Klebsiella spp. collected from intensive care units in Southern and Western Europe in 1997-1998. J Antimicrob Chemother. 2000; 45(2): 183-189.
[Crossref] [Google Scholar] [Pubmed]
- Bonomo RA, Hooper DC, Kaye KS, Johnson JR, Clancy CJ, Thaden JT, et al. Gram-negative bacterial infections: Research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis. 2017; 64(1): S30-S35.
[Crossref] [Google Scholar] [Pubmed]
- Hoban DJ, Lascols C, Nicolle LE, Badal R, Bouchillon S, Hackel M, et al. Antimicrobial susceptibility of Enterobacteriaceae, including molecular characterization of extended-spectrum beta-lactamase-producing species, in urinary tract isolates from hospitalized patients in North America and Europe: Results from the SMART study 2009-2010. Diagn Microbiol Infect Dis. 2012; 74(1): 62-67.
[Crossref] [Google Scholar] [Pubmed]
- Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, et al. CTX-M-27-and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H 30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother. 2015; 70(6): 1639-1649.
[Crossref] [Google Scholar] [Pubmed]
- Kanj SS, Kanafani ZA. Current concepts in antimicrobial therapy against resistant gram-negative organisms: Extended-spectrum ß-lactamase-producing enterobacteriaceae, carbapenem-resistant enterobacteriaceae, and multidrug-resistant Pseudomonas aeruginosa. Mayo Clin Proc. 2011; 86(3): 250-259.
[Crossref] [Google Scholar] [Pubmed]
- Nguyen HM, Shier KL, Graber CJ. Determining a clinical framework for use of cefepime and ß-lactam/ß-lactamase inhibitors in the treatment of infections caused by extended-spectrum-ß-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother. 2014; 69(4): 871-880.
[Crossref] [Google Scholar] [Pubmed]
- Harris PN, Tambyah PA, Lye DC, Mo Y, Lee TH, Yilmaz M, et al. Effect of piperacillin-tazobactam vs. meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: A randomized clinical trial. JAMA. 2018; 320(10): 984-994.
[Crossref] [Google Scholar] [Pubmed]
- Vardakas KZ, Tansarli GS, Rafailidis PI, Falagas ME. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum ß-lactamases: A systematic review and meta-analysis. J Antimicrob Chemother. 2012; 67(12): 2793-2803.
[Crossref] [Google Scholar] [Pubmed]
- Papp-Wallace KM, Kumar V, Zeiser ET, Becka SA, van den Akker F. Structural analysis of the OXA-48 carbapenemase bound to a “poor” carbapenem substrate, doripenem. Antibiotics. 2019; 8(3): 145.
[Crossref] [Google Scholar] [Pubmed]
- Kalaskar A, Venkataramana K. Determination of antimicrobial resistance pattern and production of extended-spectrum ?-lactamases amongst Escherichia coli and Klebsiella pneumoniae from clinical isolates. J Med Bacteriol. 2012; 1(3-4): 17-24.
- Ng TM, Khong WX, Harris PN, De PP, Chow A, Tambyah PA, et al. Empiric piperacillin-tazobactam versus carbapenems in the treatment of bacteraemia due to extended-spectrum beta-lactamase-producing Enterobacteriaceae. PloS One. 2016; 11(4): e0153696.
[Crossref] [Google Scholar] [Pubmed]
- Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clinical microbiology reviews. 2018; 31(2).
[Crossref] [Google Scholar] [Pubmed]
- Eftekhar F, Rastegar M, Golalipoor M, Mansoursamaei N. Detection of Extended Spectrum B-Lactamases in urinary isolates of Klebsiella pneumoniae in relation to blaSHV, blaTEM and blaCTX-M gene carriage. Iran J Public Health. 2012; 41(3): 127.
[Google Scholar] [Pubmed]
- Ranjini CY, Kasukurthi LR, Madhumati B, Rajendran R. Prevalence of multidrug resistance and extended spectrum beta-lactamases among uropathogenic Escherichia coli isolates in a tertiary care hospital in South India: An alarming trend. Community Acquired Infection. 2015; 2(1): 19.
- Sucher AJ, Chahine EB, Cogan P, Fete M. Ceftolozane/tazobactam: A new cephalosporin and ß-lactamase inhibitor combination. Ann Pharmacother. 2015; 49(9): 1046-1056.
[Crossref] [Google Scholar] [Pubmed]
- Scott LJ. Ceftolozane/tazobactam: A review in complicated intra-abdominal and urinary tract infections. Drugs. 2016; 76(2): 231-242.
[Crossref] [Google Scholar] [Pubmed]
- Solomkin J, Hershberger E, Miller B, Popejoy M, Friedland I, Steenbergen J, et al. Ceftolozane/tazobactam plus metronidazole for complicated intra-abdominal infections in an era of multidrug resistance: Results from a randomized, double-blind, phase 3 trial (ASPECT-cIAI). Clin Infect Dis. 2015; 60(10): 1462-1471.
[Crossref] [Google Scholar] [Pubmed]
- Lucasti C, Hershberger E, Miller B, Yankelev S, Steenbergen J, Friedland I, et al. Multicenter, double-blind, randomized, phase II trial to assess the safety and efficacy of ceftolozane-tazobactam plus metronidazole compared with meropenem in adult patients with complicated intra-abdominal infections. Antimicrob Agents Chemother. 2014; 58(9): 5350-5357.
[Crossref] [Google Scholar] [Pubmed]
- Seo YB, Lee J, Kim YK, Lee SS, Lee J, Kim HY, et al. Randomized controlled trial of piperacillin-tazobactam, cefepime and ertapenem for the treatment of urinary tract infection caused by extended-spectrum beta-lactamase-producing Escherichia coli. BMC Infect Dis. 2017; 17(1): 1-9.
[Crossref] [Google Scholar] [Pubmed]
- Ko JH, Lee NR, Joo EJ, Moon SY, Choi JK, Park DA, et al. Appropriate non-carbapenems are not inferior to carbapenems as initial empirical therapy for bacteremia caused by extended-spectrum beta-lactamase-producing Enterobacteriaceae: A propensity score weighted multicenter cohort study. Eur J Clin Microbiol Infect Dis. 2018; 37(2): 305-311.
[Crossref] [Google Scholar] [Pubmed]
- Pilmis B, Jullien V, Tabah A, Zahar JR, Brun-Buisson C. Piperacillin-tazobactam as alternative to carbapenems for ICU patients. Ann Intensive Care. 2017; 7(1): 1-7.
[Crossref] [Google Scholar] [Pubmed]
- Calbo E, Romaní V, Xercavins M, Gómez L, Vidal CG, Quintana S, et al. Risk factors for community-onset urinary tract infections due to Escherichia coli harbouring extended-spectrum ß-lactamases. J Antimicrob Chemother. 2006; 57(4): 780-783.
[Crossref] [Google Scholar] [Pubmed]
- Cars O, Högberg LD, Murray M, Nordberg O, Sivaraman S, Lundborg CS, et al. Meeting the challenge of antibiotic resistance. BMJ. 2008; 337.
[Crossref] [Google Scholar] [Pubmed]
- Grigoryan L, Burgerhof JG, Degener JE, Deschepper R, Lundborg CS, Monnet DL, et al. Determinants of self-medication with antibiotics in Europe: The impact of beliefs, country wealth and the healthcare system. J Antimicrob Chemother. 2008; 61(5): 1172-1179.
[Crossref] [Google Scholar] [Pubmed]
- Goossens H, Ferech M, Vander Stichele R, Elseviers M, ESAC Project Group. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet. 2005; 365(9459): 579-587.
[Crossref] [Google Scholar] [Pubmed]
- Ahmed OB, Omar AO, Asghar AH, Elhassan MM, Al-Munawwarah AM, Arabia S. Prevalence of TEM, SHV and CTX-M genes in Escherichia coli and Klebsiella spp Urinary Isolates from Sudan with confirmed ESBL phenotype. Life Sci J. 2013; 10(2): 191-195.
- Valenza G, Nickel S, Pfeifer Y, Eller C, Krupa E, Lehner-Reindl V, et al. Extended-spectrum-ß-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob Agents Chemother. 2014; 58(2): 1228-1230.
[Crossref] [Google Scholar] [Pubmed]
- Lonchel CM, Melin P, Gangoue´-Pie´boji J, Assoumou MC, Boreux R, de Mol P. Extended-spectrum ß-lactamase-producing Enterobacteriaceae in Cameroonian hospitals. Eur J Clin Microbiol Infect Dis. 2013; 32(1): 79-87.
[Crossref] [Google Scholar] [Pubmed]
- Bonnet R. Growing group of extended-spectrum ß-lactamases: The CTX-M enzymes. Antimicrob Agents Chemother. 2004; 48(1): 1-4.
[Crossref] [Google Scholar] [Pubmed]
- Wiener J, Quinn JP, Bradford PA, Goering RV, Nathan C, Bush K, et al. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA. 1999; 281(6): 517-523.
[Crossref] [Google Scholar] [Pubmed]
- Shahid M, Singh A, Sobia F, Rashid M, Malik A, Shukla I, et al. blaCTX-M, blaTEM, and blaSHV in Enterobacteriaceae from North-Indian tertiary hospital: High occurrence of combination genes. Asian Pac J Trop Med. 2011; 4(2): 101-105.
[Crossref] [Google Scholar] [Pubmed]
- Moses A, Bwanga F, Boum Y, Bazira J. Prevalence and genotypic characterization of Extended-Spectrum Beta-Lactamases produced by gram negative bacilli at a tertiary Care Hospital in Rural South Western Uganda. Br Microbiol Res J. 2014; 4(12): 1541.
[Crossref] [Google Scholar] [Pubmed]
- Tamang MD, Nam HM, Gurung M, Jang GC, Kim SR, Jung SC, et al. Molecular characterization of CTX-M ß-lactamase and associated addiction systems in Escherichia coli circulating among cattle, farm workers, and the farm environment. Appl Environ Microbiol. 2013; 79(13): 3898-3905.
[Crossref] [Google Scholar] [Pubmed
Author Info
Ivan Muhwezi1*, Joel Bazira1, Henry Zamarano1, Frederick Byarugaba1, Wilber Sabiiti2, Mathew Holden2 and Benon B Asiimwe32Department of Medicine, University of St Andrews, St Andrews, United Kingdom
3Department of Medical Microbiology, School of Biomedical Sciences, College of Health Sciences, Makerere University,, Kampala, Uganda
Citation: Muhwezi I: Quantification and Molecular Characterization of Extended Spectrum Beta-Lactamase Producing Enterobacteriaceae from Agropastoral Communities of Mbarara District, South Western Uganda
Received: 20-Oct-2022 Accepted: 03-Nov-2022 Published: 10-Nov-2022, DOI: 10.31858/0975-8453.13.11.733-743
Copyright: This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
ARTICLE TOOLS
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Psychometric properties of the World Health Organization Quality of life instrument, short form: Validity in the Vietnamese healthcare context Trung Quang Vo*, Bao Tran Thuy Tran, Ngan Thuy Nguyen, Tram ThiHuyen Nguyen, Thuy Phan Chung Tran SRP. 2020; 11(1): 14-22 » doi: 10.5530/srp.2019.1.3
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- Deuterium Depleted Water as an Adjuvant in Treatment of Cancer Anton Syroeshkin, Olga Levitskaya, Elena Uspenskaya, Tatiana Pleteneva, Daria Romaykina, Daria Ermakova SRP. 2019; 10(1): 112-117 » doi: 10.5530/srp.2019.1.19
- Dental Development between Assisted Reproductive Therapy (Art) and Natural Conceived Children: A Comparative Pilot Study Norzaiti Mohd Kenali, Naimah Hasanah Mohd Fathil, Norbasyirah Bohari, Ahmad Faisal Ismail, Roszaman Ramli SRP. 2020; 11(1): 01-06 » doi: 10.5530/srp.2020.1.01
- Manilkara zapota (L.) Royen Fruit Peel: A Phytochemical and Pharmacological Review Karle Pravin P, Dhawale Shashikant C SRP. 2019; 10(1): 11-14 » doi: 0.5530/srp.2019.1.2
- Pharmacognostic and Phytopharmacological Overview on Bombax ceiba Pankaj Haribhau Chaudhary, Mukund Ganeshrao Tawar SRP. 2019; 10(1): 20-25 » doi: 10.5530/srp.2019.1.4
- A Review of Pharmacoeconomics: the key to “Healthcare for All” Hasamnis AA, Patil SS, Shaik Imam, Narendiran K SRP. 2019; 10(1): s40-s42 » doi: 10.5530/srp.2019.1s.21
- A Prospective Review on Phyto-Pharmacological Aspects of Andrographis paniculata Govindraj Akilandeswari, Arumugam Vijaya Anand, Palanisamy Sampathkumar, Puthamohan Vinayaga Moorthi, Basavaraju Preethi SRP. 2019; 10(1): 15-19 » doi: 10.5530/srp.2019.1.3